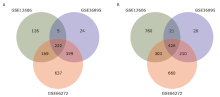
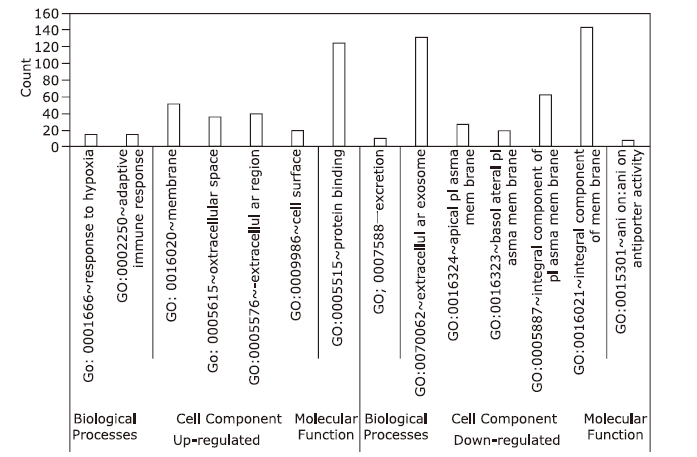
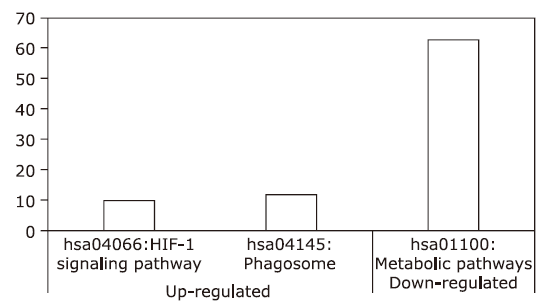
| [1.] |
Li P, Znaor A, Holcatova I, et al. Regional geographic variations in kidney cancer incidence tates in European countries. Eur Urol 2015; 67(6):1134-41. doi: 10.1016/j.eururo.2014.11.001.
doi: 10.1016/j.eururo.2014.11.001
|
| [2.] |
Srinivasan R, Ricketts CJ, Sourbier C, et al. New strategies in renal cell carcinoma: targeting the genetic and metabolic basis of disease. Clin Cancer Res 2015; 21(1):10-7. doi: 10.1158/1078-0432.CCR-13-2993.
doi: 10.1158/1078-0432.CCR-13-2993
|
| [3.] |
Siegel R, Naishadham D and Jemal A. Cancer statistics. CA Cancer J Clin 2013; 63(1):11-30. doi: 10.3322/caac.21166.
doi: 10.3322/caac.21166
|
| [4.] |
Capitanio U and Montorsi F. Renal cancer. Lancet 2016; 387(10021):894-906. doi: 10.1016/S0140-6736(15)00046-X.
doi: 10.1016/S0140-6736(15)00046-X
pmid: 26318520
|
| [5.] |
Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013; 339(6127):1546-58. doi: 10.1126/science.1235122.
doi: 10.1126/science.1235122
|
| [6.] |
Feng H, Gu ZY, Li Q, et al. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis . J Ovarian Res 2019; 12(1):35. doi: 10.1186/s13048-019-0508-2.
doi: 10.1186/s13048-019-0508-2
pmid: 31010415
|
| [7.] |
Na YQ, Ye ZQ, Sun YH, et al. Diagnosis and treatment of renal cell carcinoma guide manual. In: Xiao J, Yand F, editors. The Chinese department of urology disease diagnosis and treatment guide manual, 2014 ed. Beijing: People Health Publishing House, 2013. p. 1-29.
|
| [8.] |
Li DY, Song YS. Comparison of clinical data of renal cell carcinoma between elderly and middle aged and young patients. J Modem Oncology 2017; 25(5):740-3. doi: 10.3969/j. issn.1672-4992.2017.05.018.
doi: 10.3969/j. issn.1672-4992.2017.05.018
|
| [9.] |
Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol 2003; 170(6 pt 1):2163-72. doi: 10.1097/01.ju.0000096060.92 397.ed.
doi: 10.1097/01.ju.0000096060.92 397.ed
|
| [10.] |
Duensing S, Hohenfellner M. Adjuvant therapy for renal cell carcinoma: settled for now. Lancet 2016; 387(10032):1973-4. doi: 10.1016/S0140-6736(16)00653-X.
doi: 10.1016/S0140-6736(16)00653-X
pmid: 26969091
|
| [11.] |
Krabbe LM, Bagrodia A, Margnlis V, et al. Surgical management of renal cell carcinoma. Semin Intervent Radiol 2014; 31(1):27-32. doi: 10.1055/s-0033-1363840.
doi: 10.1055/s-0033-1363840
|
| [12.] |
Kim HS, Park KH, Sun AK, et al. Frequent mutations of human Mad2, but not Bub1, in gastric cancers cause defective mitotic spindle checkpoint. Mutat Res 2005; 578(1-2):187-201. doi: 10.1016/j. mrfmmm.2005.05.020.
doi: 10.1016/j. mrfmmm.2005.05.020
|
| [13.] |
Mur P, De Voer RM, Olivera-Salguero R, et al. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are infrequent in familial colorectal cancer and polyposis. Mol Cancer 2018; 17(1):23. doi: 10.1186/s12943-018-0762-8.
doi: 10.1186/s12943-018-0762-8
|
| [14.] |
Myrie KA, Percy MJ, Azim JN, et al. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett 2000; 152(2):193-9. doi: 10.1016/s0304-3835(00)00340-2.
doi: 10.1016/s0304-3835(00)00340-2
pmid: 10773412
|
| [15.] |
Burum-Auensen E, Deangelis PM, Schjølberg AR, et al. Spindle proteins Aurora A and BUB1B, but not Mad2, are aberrantly expressed in dysplastic mucosa of patients with longstanding ulcerative colitis. J Clin Pathol 2017; 60(12):1403-8. doi: 10.1136/jcp.2006.044305.
doi: 10.1136/jcp.2006.044305
|
| [16.] |
Moustakas A. The mitotic checkpoint protein kinase BUB1 is an engine in the TGF-β signaling apparatus. Sci Signal 2015; 8(359):fs1. doi: 10.1126/scisignal.aaa4636.
doi: 10.1126/scisignal.aaa4636
|
| [17.] |
Nyati S, Schinske-sebolt K, Pitchiaya S, et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-β signaling. Sci Signal 2015; 8(358):ra1. doi: 10.1126/scisignal.2005379.
doi: 10.1126/scisignal.2005379
|
| [18.] |
Qian XT, Song XK, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 2015; 74:222-7. doi: 10.1016/j.biopha.2015.08.004.
doi: 10.1016/j.biopha.2015.08.004
|
| [19.] |
Vermeulen K, Bockstaele DRV, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 2003; 36(3):131-49. doi: 10.1046/j.1365-2184.2003.00266.x.
doi: 10.1046/j.1365-2184.2003.00266.x
|
| [20.] |
Park SH, Yu GR, Kim WH, et al. NF-Y-dependent cyclin B2 expression in colorectal adenocarcinoma. Clin Cancer Res 2007; 13(3):858-67. doi: 10.1158/1078-0432.CCR-06-1461.
doi: 10.1158/1078-0432.CCR-06-1461
|
| [21.] |
Wang DG, Chen G, Wen XY, et al. Identification of biomarkers for diagnosis of gastric cancer by bioinformatics. Asian Pac J Cancer Prev 2015; 16(4):1361-5. doi: 10.7314/apjcp.2015.16.4.1361.
doi: 10.7314/apjcp.2015.16.4.1361
|
| [22.] |
Mo ML, Chen Z, Li J, et al. Use of serum circulating Ccnb2 in cancer surveillance. Int J Biol Markers 2010; 25(4):236-42. doi: 10.5301/JBM.2010.6088.
doi: 10.5301/JBM.2010.6088
|
| [23.] |
Yamamura S, Kawakami K, Hirata H, et al. Oncogenic functions of secreted frizzled-related protein 2 in human renal cancer. Mol Cancer Ther 2010; 9(6):1680-7. doi: 10.1158/1535-7163.MCT-10-0012.
doi: 10.1158/1535-7163.MCT-10-0012
|
| [24.] |
Hong Y, Huang X, An L, et al. Overexpression of COPS3 promotes clear cell renal cell carcinoma progression viaregulation of phospho-AKT (Thr308), cyclin D1 and caspase-3 . Exp Cell Res 2018; 365(2):163-70. doi: 10.1016/j.yexcr.2018.02.025.
doi: 10.1016/j.yexcr.2018.02.025
|
| [25.] |
Haddad AQ, Luo JH, Krabbe LM, et al. Prognostic value of tissue-based biomarker signature in clear cell renal cell carcinoma. BJU Int 2017; 119(5):741-7. doi: 10.1111/bju.13776.
doi: 10.1111/bju.13776
|
| [26.] |
Wei C, Yang XL, Xi JH, et al. High expression of pituitary tumor-transforming gene-1 predicts poor prognosis in clear cell renal cell carcinoma. Mol Clin Oncol 2015; 3(2):387-91. doi: 10.3892/mco.2014.478.
doi: 10.3892/mco.2014.478
|
| [27.] |
Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol 1997; 11(4):433-41. doi: 10.1210/mend.11.4.9911.
doi: 10.1210/mend.11.4.9911
pmid: 9092795
|
| [28.] |
Feng ZZ, Chen JW, Yang ZR, et al. Expression of PTTG1 and PTEN in endometrial carcinoma: correlation with tumorigenesis and progression. Med Oncol 2012; 29(1):304-10. doi: 10.1007/s12032-010-9775-x.
doi: 10.1007/s12032-010-9775-x
|
| [29.] |
Yoon CH, Kim MJ, Lee H, et al. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population . J Biol Chem 2012; 287(23):19516-27. doi: 10.1074/jbc.M111.337428.
doi: 10.1074/jbc.M111.337428
|
| [30.] |
Rahman MA, Amin ARMR, Wang X, et al. Systemic delivery of siRNA nanoparticles targeting RRM2 suppresses head and neck tumor growth. J Control Release 2012; 159(3):384-92. doi: 10.1016/j.jconrel.2012.01.045.
doi: 10.1016/j.jconrel.2012.01.045
|
| [31.] |
Morikawa T, Hino R, Uozaki H, et al. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibitionin vitro . Hum Pathol 2010; 41(12):1742-8. doi: 10.1016/j.humpath.2010.06.001.
doi: 10.1016/j.humpath.2010.06.001
pmid: 20825972
|
 )
)
 )
)