Chinese Medical Sciences Journal ›› 2017, Vol. 32 ›› Issue (3): 135-144.doi: 10.24920/J1001-9294.2017.032
• Original Article • Next Articles
Genetic Correction and Hepatic Differentiation of Hemophilia B-specific Human Induced Pluripotent Stem Cells
He Qiong1, Wang Hui-hui1, 2, Cheng Tao3, Yuan Wei-ping3, Ma Yu-po4, 5, 6, *( ), Jiang Yong-ping1, Ren Zhi-hua1, 2, *(
), Jiang Yong-ping1, Ren Zhi-hua1, 2, *( )
)
- 1Biopharmaceutical R&D Center, Chinese Academy of Medical Sciences & Peking Union Medical College, Suzhou 215126, China;
2 Biopharmagen Corporation, Suzhou 215126, China
3 State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, Center for Stem Cell Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin 300020, China;
4 iCell Gene Therapeutics LLC, Research & Development Division, Long Island High Technology Incubator, Stony Brook, NY 11794, USA;
5 Department of Pathology, Stony Brook Medicine, Stony Brook, NY 11794, USA
6 Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau 999078, China
-
Received:2017-03-16Published:2017-09-27Online:2017-09-27 -
Contact:Ma Yu-po,Ren Zhi-hua E-mail:yupo.ma@stonybrookmedicine.edu;renzh@biopharmagen.com
Cite this article
He Qiong, Wang Hui-hui, Cheng Tao, Yuan Wei-ping, Ma Yu-po, Jiang Yong-ping, Ren Zhi-hua. Genetic Correction and Hepatic Differentiation of Hemophilia B-specific Human Induced Pluripotent Stem Cells[J].Chinese Medical Sciences Journal, 2017, 32(3): 135-144.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primers for disease-causing point mutation screening"
| Exons | Primer sequences | Product length (bp) |
|---|---|---|
| Exon 1 | Forward: CCACTGCCCATTCTCTTCA Reverse: TACTTACCAACCTGCGTGCT | 393 |
| Exon 2+3 | Forward: CATGCCCTAAAGAGAAATTGG Reverse: TGGGTTAGAGGGTTGGACTG | 639 |
| Exon 4 | Forward: TCAGGAAGACAGGAGCATCA Reverse: GAGGGAAACTTTGAACCATGAG | 322 |
| Exon 5 | Forward: ACCCATACATGAGTCAGTAGTTCC Reverse: GACACAGAAAGAATTCAGGTTGTAG | 427 |
| Exon 6 | Forward: AATACTGATGGGCCTGCTTC Reverse: ACCTGGCCTGTGTCTTGC | 383 |
| Exon 7 | Forward: GCCTATTCCTGTAACCAGCAC Reverse: GACCCTTCTGCCTTTAGCC | 324 |
| Exon 8 | Forward: CAATTAGGTCAGTGGTCCCAAG Reverse: GGGAAAGTGATTAGTTAGTGAGAGG | 713 |
Table 2
Primers of the top 8 theoretical off-target sites"
| No. | Off-target site | Primers |
|---|---|---|
| OT 1 | GACAGCAGTTTGGTTGTTGG GAG | Forward: gttaattcccactcctgaagaga Reverse: tggttaaaaactctcaggcagc |
| OT 2 | GCCTCCTCTTGGGTTGTTTG GAG | Forward: ttctttagatcattttatttaaaaacagtgc Reverse: aatcaccccattcaggtctaa |
| OT 3 | GGCTTCCCATGAGTTGTTGG CAG | Forward: tcctctaccacgtgggctac Reverse: agagtccaggcttgaaccac |
| OT 4 | CACTGCACTGGGGTTCTTGG GGG | Forward: cggaccccagactcttccta Reverse: aagcttccagaggacggtgg |
| OT 5 | GAGTGCACTTGGGTGGCTGG AGG | Forward:acccagtcctcacttggtta Reverse: acagccttcagtgcagtcct |
| OT 6 | AGCTTCATTTGGGTTGTTGG AAG | Forward: actcacctaacaaaagctctttcaa Reverse: tccctgcctccctgtcttta |
| OT 7 | AATTTCTCTTGGGTTGTTGG GAG | Forward: tcgttatttttgtaaggctccta Reverse: aacactaatccatgagatctgaaaa |
| OT 8 | GGCTGCACTTGGGTTGTTTG AGG | Forward: tgcctactgacaacaagagca Reverse: aagggacagggtgaaagagg |
Table 3
Information about primary and secondary antibodies"
| Antigen | Distributor | Host | Cat# | Dilution |
|---|---|---|---|---|
| OCT3/4 | Santa Cruz | Rabbit | sc-9081 | 1:500 |
| SOX17 | ABCAM | Mouse | Ab84990 | 1:50-100 |
| AFP | ABCAM | Rabbit | ab169552 | 1:100-250 |
| ALB | R&D System | Mouse | MAB1455 | 8-25 μg/ml |
| F IX (F9) | Life science products & services | Rabbit | AB21319a | 1:25-100 |
| Fluorescein (FITC)-conjugated AffiniPure F(ab’) 2 Fragment Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch Laboratories | Donkey | 715-096-150 | 1:50-200 |
| CyTM3-conjugated AffiniPure F(ab’)2 Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Laboratories | Donkey | 711-166-152 | 1:100-800 |

Figure 1.
Construction of CRISPR-sgRNA plasmids and design of the 129-nt homologous repair template.A. Sequences of the 4 selected sgRNAs. B. Characteristics of the 4 selected sgRNAs. C. Cleavage activity validation of the 4 CRISPR-sgRNAs. D. Translation of sequences round the targeted locus. E. Codons of some interested amino acids. F. Sequence information of the homologous template. iPSCs: induced pluripotent stem cells. Basepairs highlighted in red indicate point mutation site or correction site; basepairs highlighted in green represent synonymous mutations; basepairs highlighted in blue indicate protospacer adjacent motif region."


Figure 3.
Sequencing chromatograph. A. Sequencing chromatograph of the patient derived iPSC clone. The arrow showed the missense point mutation. B. Sequencing chromatograph of the ten genetically corrected clones. Clone 4 and Clone 52 had two peaks at the mutation point, indicating these two clones were not pure."
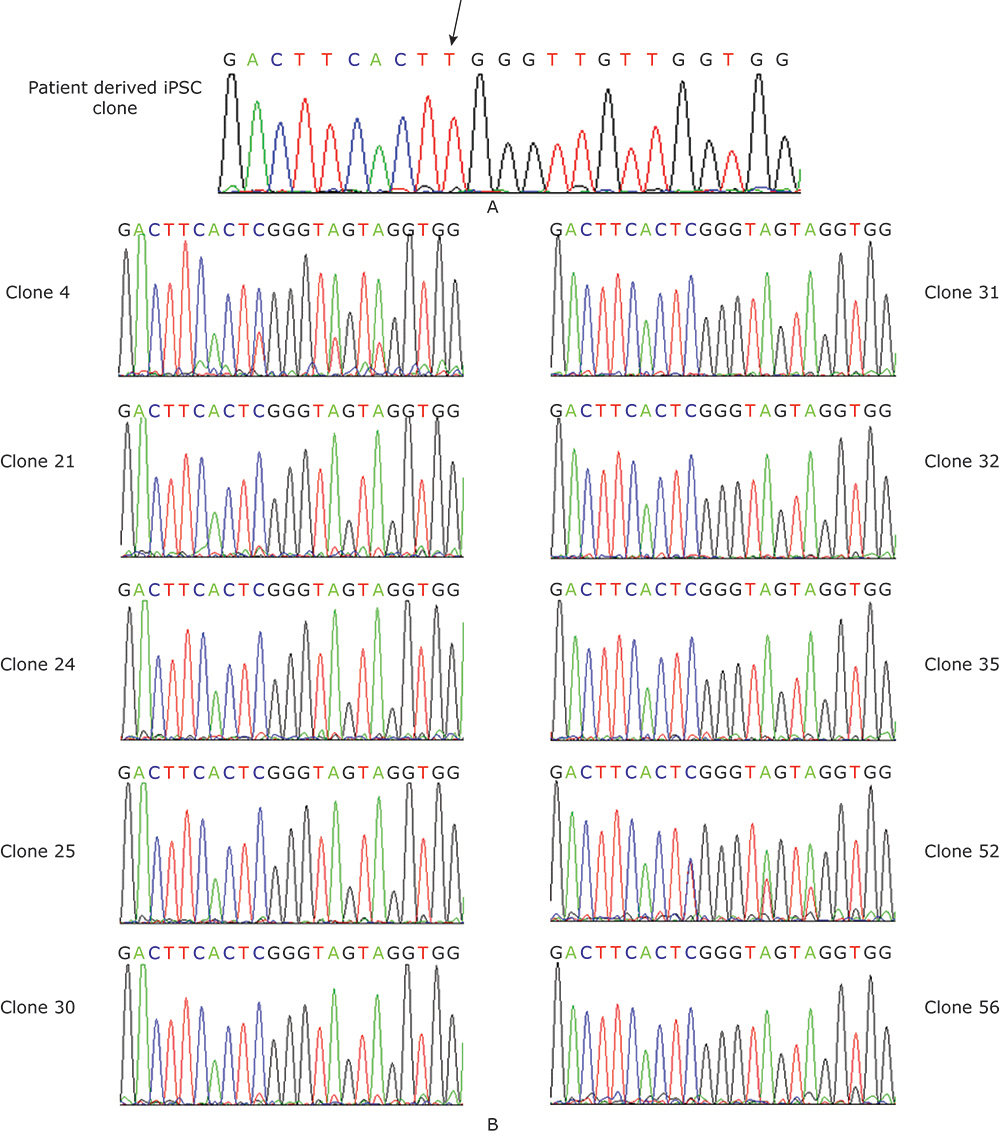

Figure 4.
Deviation of hepatocyte-like cells from genetically corrected iPSCs.A, B, C. Compact colony morphologies, with a high ratio of nucleus to cytoplasm and prominent nucleoli, were observed. Undifferentiated iPSCs were identified by immunocytochemistry using antibody that recognized OCT3/4. D, E, F. Characteristics of hepatic differentiation procedure in vitro. Expression of SOX17, AFP and ALB were detected on day 5, day 15 and day 20, respectively. Scale bar = 50 μm."


Figure 5.
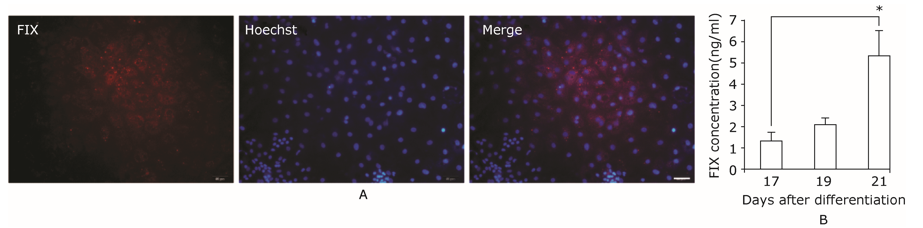
Factor IX expressed in hepatic differentiated cells.A. Coagulation factor IX expressed in hepatic cells derived from iPSCs. F IX was located in cytoplasm and further secreted to culture medium. Scale bar = 50 μm. B. Quantification of FIX released in culture medium by ELISA assay. *indicates P≤0.01."
| 1. | Chen Y, Schroeder JA, Kuether EL, Zhang G, Shi Q.Platelet gene therapy by lentiviral gene delivery to hematopoietic stem cells restores hemostasis and induces humoral immune tolerance in FIX(null) mice. Mol Ther 2014; 22(1):169-77. doi: 10.1038/mt.2013.197. |
| 2. | Li T, Miller CH, Driggers J, Payne AB, Elliingsen D, Hooper WC.Mutation analysis of a cohort of US patients with hemophilia B. Am J Hematol 2014; 89(4):375-9. doi: 10.1002/ajh.23645. |
| 3. | Bowen DJ.Haemophilia A and haemophilia B: molecular insights. Mol Pathol 2002; 55(1):1-18. doi: 10.1136/mp. 55.1.1. |
| 4. | Seo JY, Jang MA, Kim HJ, Lee KO, Kim SH, Kim HJ.Sequence variation data of F8 and F9 genes in functionally validated control individuals: implications on the molecular diagnosis of hemophilia. Blood Res 2013; 48(3): 206-10. doi: 10.5045/br.2013.48.3.206. |
| 5. | Takahashi K, Yamanaka S.Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126(4):663-76. doi: 10.1016/ j.cell.2006.07.024. |
| 6. | Takahashi K, Okita K, Nakagawa M, Yamanaka S.Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2007; 2(12):3081-9. doi: 10.1038/nprot.2007. 418. |
| 7. | Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif 2008; 41 Suppl 1:51-6. doi: 10.1111/j.1365-2184.2008. 00493.x. |
| 8. | Iwamuro M, Komaki T, Kubota Y, Seita M, Kawamoto H, Yuasa T, et al.Comparative analysis of endoderm formation efficiency between mouse ES cells and iPS cells. Cell Transplant 2010; 19(6):831-9. doi: 10.3727/096368 910X508951. |
| 9. | Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, et al.Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res 2009; 19(11):1233-42. doi: 10.1038/cr.2009.107. |
| 10. | Paques F, Duchateau P.Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther 2007; 7(1):49-66. doi: 10.2174/156652307779940216. |
| 11. | Carroll D.Genome engineering with zinc-finger nucleases. Genetics 2011; 188(4):773-82. doi: 10.1534/genetics. 111.131433. |
| 12. | Wright DA, Li T, Yang B, Spalding MH.TALEN-mediated genome editing: prospects and perspectives. Biochem J 2014; 462(1):15-24. doi: 10.1042/BJ20140295. |
| 13. | Doudna JA, Charpentier E.Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346(6213):1258096. doi: 10.1126/science. 1258096. |
| 14. | Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F.Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8(11):2281-308. doi: 10.1038/nprot.2013. 143. |
| 15. | Hsu PD, Lander ES, Zhang F.Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157(6):1262-78. doi: 10.1016/j.cell.2014.05.010. |
| 16. | Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, et al.Highly efficient generation of human hepatocyte- like cells from induced pluripotent stem cells. Hepatology 2010; 51(1):297-305. doi: 10.1002/hep.23354. |
| 17. | Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J.RNA-programmed genome editing in human cells. Elife 2013; 2:e00471. doi: 10.7554/eLife.00471. |
| 18. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337(6096):816-21. doi: 10.1126/science.1225829. |
| 19. | Cone L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al.Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339(6121):819-23. doi: 10.1126/ science.1231143. |
| 20. | Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science 2013; 339(6121):823-6. doi: 10.1126/science. 1232033. |
| 21. | Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016; 351(6271): 407-11. doi: 10.1126/science.aad5177. |
| 22. | Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al.Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014; 32(6):551-3. doi: 10.1038/nbt.2884. |
| 23. | Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al.Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res 2014; 115(5): 488-92. doi: 10.1161/CIRCRESAHA.115.304351. |
| 24. | Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, et al.The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids 2014; 3:e186. doi: 10.1038/mtna.2014.38. |
| 25. | Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, et al.CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med 2016; 8(5):477-88. doi: 10.15252/emmm.201506039. |
| 26. | Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, et al.Seamless gene correction of b-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 2014; 24(9):1526-33. doi: 10.1101/gr. 173427.114. |
| 27. | Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al.Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cysticfibrosis patients. Cell Stem Cell 2013; 13(6):653-8. doi: 10.1016/j.stem.2013.11.002. |
| 28. | Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, et al.Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep 2015; 4(1):143-54. doi: 10.1016/j.stemcr.2014. 10.013. |
| 29. | Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA.Multiplex CRISPR/Cas9-based genome editing forcorrectionofdystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 2015; 6:6244. doi: 10.1038/ncomms7244. |
| 30. | Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al.High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013; 31(9):822-6. doi: 10.1038/nbt.2623. |
| 31. | Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al.DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31(9):827-32. doi: 10.1038/nbt.2647. |
| 32. | Wu XB, Andrea JK, Philip AS.Target specificity of the CRISPR-Cas9 system. Quant Biol 2014, 2(2):59-70. doi: 10.1007/s40484-014-0030-x. |
| 33. | Ashrani AA, Reding MT, Shet A, Osip J, Humar A, Lake JR, et al.Successful liver transplantation in a patient with severe haemophilia A and a high-titre factor Ⅷ inhibitor. Haemophilia 2004; 10(6):735-7. doi: 10.1111/j.1365-2516. 2004.01030.x. |
| 34. | Wilde J, Teixeira P, Bramhall SR, Gunson B, Mutimer D, Mirza DF.Liver transplantation in haemophilia. Br J Haematol 2002; 117(4):952-6. doi: 10.1046/j.1365-2141. 2002.03528.x. |
| 35. | Gordon FH, Mistry PK, Sabin CA, Lee CA.Outcome of orthotopic liver transplantation in patients with haemophilia. Gut 1998; 42(5):744-9. doi: 10.1136/gut.42.5.744. |
| 36. | Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 2014; 371(21):1994-2004. doi: 10.1056/NEJMoa 1407309. |
| 37. | Sharma A, Easow MM, Sriganesh V, Neely JA, Kalipatnapu S. Gene therapy for haemophilia. Cochrane Database Syst Rev 2014; 11:CD010822. doi: 10.1002/ 14651858.CD010822.pub2. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
|



