CONVENTIONAL disease diagnostics rely on iconography or antibody-based platform which are slow and costly.1 Synthetic biology which enables cells to perform designed novel tasks by modifying the native gene pathways has provided an alternative approach for disease detection. A gene pathway can be regarded as a network consisting of a series of functional gene modules. Synthetic biologists reprogram cells to perform designed tasks by integrating different functional gene modules into cells. This promising technology has been successfully utilized to reprogram a wide range of different cell species from prokaryotes like Escherichia coli and lower eukaryotes like yeast to higher eukaryotes like mammalian cells, to perform novel tasks with great diversity, such as producing new chemicals and proteins, drug discovery. In this review, we give an overview of recent advances in the application of synthetic biosensors for decease detection. The design principles and characteristics of synthetic biosensors with different architectures are summarized.
DESIGN PRINCIPLES OF SYNTHETIC BIOSENSORS
An ideal synthetic biosensor for disease detection is reminiscent of a precise regulation system. It consists of two functional modules: the detecting module and the responding module. The detecting module precisely detects biomarker signals and triggers the responding module to express desired reporters. Biology enriches with plenty of perfect gene regulation systems. The first regulation system, lac operon in Escherichia coli was discovered by Jacob and Monod in 1961.2 The lac operon exhibits a subtle regulatory control of lactose metabolism. There are three structural genes of lac operon, lacZ, lacY, and lacA. They encode enzymes necessary for lactose catabolism. In the absence of lactose, lac repressor which is constitutively expressed in E.coli binds to the DNA sequence, called lac operator, downstream of the promoter and inhibits the transcription of lacZ, lacY, and lacA and consequently halts the production of the enzymes encoded. In the presence of lactose, on the contrary, a lactose metabolite called allolactose, binds to the lac repressor, causing an allosteric shift. Then the repressor is released from the DNA sequence, allowing the transcription of lac genes and thereby leading to higher levels of the enzymes. The lac operon shows how biology control cellular metabolism in an efficient, precise and energy saving manner.
Since the discovery of the lac operon, more and more regulation systems have been discovered and identified, most of which are in prokaryotes.3 Inspired by electronic circuits, synthetic biologists have integrated different functional parts of these regulation systems into gene circuits which act in a biosensor manner. These heterologous synthetic biosensors enabled cells precisely and efficiently respond to signals from their microenvironment. A synthetic biosensor for disease detection is composed of three key elements: a signal, a transcription factor, and an operator.
In the disease detection manner, a signal is the biomarker representing the pathological conditions of the cell and/or system. It triggers the binding or releasing of a transcription factor from the operator, which then controls the expression of the reporter. To date, a great number of prokaryotic transcription factors have been identified that respond to a variety of signals including antibiotics,4-6 amino acids,7 food additives,8 metabolites, hormones, vitamins, cell-cell signaling molecules9-11 and metals.12
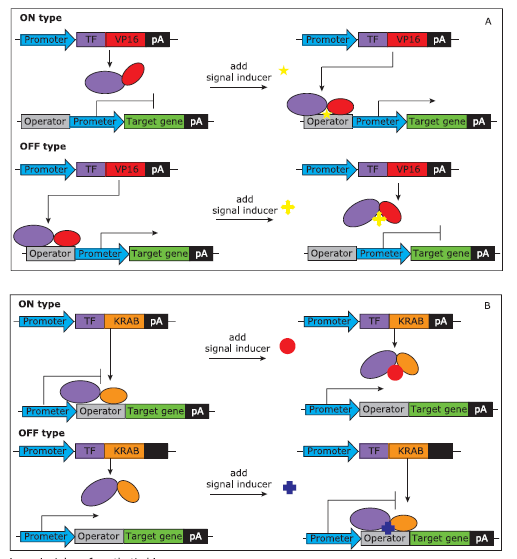
Transcription factors can be classified into two distinct classes: the activator and the repressor. In the activation manner, prokaryotic transcription factors are fused to a transactivation domain Herpes simplex virus virion protein 16 (VP16),13 forming a functional mammalian chimeric transcription factor. It acts as a transcription activator of target genes by binding to the operator of the promoter. The DNA binding activity of the chimeric transcription factor can be activated or repressed by signals according to different allosteric shifts, therefore forming an ON type or an OFF type system (Fig. 1A). In the repression manner, prokaryo-tic regulators are fused to transcription-silencing domain Krueppel associated box (KRAB) to repress the transcription of target genes when bound to operators located in the vicinity of the promoter. The DNA binding of the chimeric transcription factor can be active or repressed by the signal according to different allosteric shifts, therefore constructing an OFF type or an ON type system (Fig. 1B).
An operator is a DNA sequence that the transcription factor can bind to, which will modulate the activity of the target gene. An operator can be composed of a single copy or multiple tandem copies of the minimal DNA-binding sequence and can be located either upstream and/or downstream of the promoter.
A. Activation-based ON and OFF synthetic biosensor. A prokaryotic transcription factor is fused to a transactivation domain, Herpes simplex virus protein 16 (VP16) , forming a functional mammalian chimeric transcription factor. It acts as a transcription activator of target genes by binding to the operator of the promoter. In an ON-type synthetic biosensor, the signal inducer triggers the binding of the chimeric transcription factor to the operator thus turn on the transcription of the target gene. In an OFF-type synthetic biosensor, the signal inducer represses the binding of the chimeric transcription factor to the operator thus turn off the transcription of the target gene.
B. Repression-based ON and OFF synthetic biosensor. A prokaryotic regulator is fused to transcription-silencing domain Krueppel associated box(KRAB) to repress the transcription of target genes when bound to operators located in the vicinity of the promoter. In an ON-type synthetic biosensor, the signal inducer represses the binding of chimeric transcription factor to the operator thus turn on the transcription of the target gene. In an OFF-type synthetic biosensor, the signal inducer triggers the binding of the chimeric transcription factor to the operator thus turn off the transcription of the target gene.
SIMPLE SYNTHETIC BIOSENSORS
Simple synthetic biosensors are the basic biosensor device composed of a signal sensing module and a responding module. The tetracycline (Tet) on/off synthetic biosensor is the most commonly used one. It is derived from the prokaryotic tetracycline repressor (TetR) transcription system. TetR transcriptional repressors constitute one of the most abundant transcription family of prokaryotic regulators.3,12 It responds to antibiotic tetracycline and the homologous. In the absence of tetracycline, TetR forms a dimer that strongly binds to the TetR operator sequence (tetO). In the presence of tetracycline, the dimer is disrupted, TetR dissociates from the DNA, and gene expression is activated.
In a synthetic biosensor, TetR can be constructed as either a repressor, or converted into an activator by fusing it to the transactivation domain VP16. Single to multiple copies of tetO are placed upstream of the promoter, referred to as a Tetracycline Response Element or TRE. In a “Tet-Off” system, the reporter expression is activated when TetR is bound to the TRE, and becomes inactivated upon the addition of doxycycline which realizes the binding of TetR to TRE. In a “Tet-On” system, the TetR transcription factor is reversely fused to VP16, referred to as rtTA. In this fashion, the reporter expression is activated upon addition of doxycycline.14 These synthetic biosensors typically exhibit a large dynamic range (from 10 to several thousand-fold induction) and have been shown to function in a wide range of cell lines, including embryonic stem cells,15,16 CHO,17,HEK,18 HeLa19 and MCF-720 cells.
During the past decade, synthetic biosensors have been rapidly developed from simple control devices to multi-gene/protein-based transcription and signaling networks, from linear manner regulation to looped manner regulation. These developments not only broadened the application of biosensors, but also improved the performance of biosensors. Leakiness was the typical problem of the early generation biosensor. A series of studies focused on this issue by investigating different promotes, optimizing the number of operator modules, the relative spacing of the operator modules to the promoter, and the resulting torsion angle of the operator-bound transactivator in order to eliminate the leakiness. Side effects was another issue of the early generation biosensor because most trigger molecules, e.g. antibiotics, hormones, immunosuppressive drugs exhibit therapeutic side effects. To overcome this problem, adjustable transgene expression circuits controlled by endogenous physiologic metabolites, food derived compounds, vitamins, or pathologic signals have been gradually designed.21 With more regulation system identified, a series of synthetic biosensors responding to various signals have been built based on similar design principles of the Tet on/off system. Table 1 summarizes detail characteristics of various simple synthetic biosensors.
Table 1 Characteristics of various simple synthetic biosensors
| Classification of signal | On/Off | Signals | Regulation factor | Regulation factor fused domain | Chimeric regulation factor | Applications | Reference |
|---|---|---|---|---|---|---|---|
| Antibiotic | |||||||
| On and Off | Macrolide (erythromycin, clarithromycin, and roxithromycin) | MphR(A) | VP16 and KRAB | ET | Gene therapy, tissue engineering, in vivo gene function analyses, drug discovery, biopharmaceutical manufacturing | 5 | |
| On and Off | Pristinamycin I | Pip | VP16 and KRAB | PIT | Compatible with the Tet-OFF system, thus two different gene activities can be controlled in the same cell | 6 | |
| Amino acid | |||||||
| On | L-arginine | ArgR | ART | VP16 | Gene therapy and manufacturing of protein pharmaceuticals | 7 | |
| On | Tryptophan | TrpR | VP16 | TRT | Targeted gene expression control | 57 | |
| Small molecular | |||||||
| On and Off | Cumate | CymR | VP16 | cTA, rcTA | Regulation of gene expression level and duration | 58 | |
| Gas | |||||||
| On | Acetaldehyde | AlcR | - | AlcR | Therapeutic transgene dosing and biopharmaceutical manufacturing | 59 | |
| On | 6-hydroxy-nicotine | HdnoR | VP16 | - | Drug-responsive homologs in basic research, therapeutic cell engineering and biopharmaceutical manufacturing. | 60 | |
| On | pH/CO2 | TDAG8 | - | - | Remote control of cellular behavior inside microfluidic devices; CO2-triggered production of biopharmaceuticals in standard bioreactors | 61 | |
| Drug compound | |||||||
| On | 2-phenylethyl-butyrate | EthR | VP16 | - | Generic screening platform to discover drug candidates | 62 | |
| Qutam sensing signal | |||||||
| On | Butyrolactones | ScbR SpbR | VP16 | SCA, SPA | Clinical application | 9 | |
| On | 3-oxo-C8-HSL | TraR | NF-κB p65 | p65NTraR | Versatile gene expression control | 10 | |
| Metabolites | |||||||
| On | Uric acid | HucR | KRAB | mUTS | Self-sufficient control of pathologic metabolites, gene- and cell-based therapies | 54 | |
| On | Bile acids | TGR5 | - | p65NTraR | Liver injuries protection | 44 | |
| On | Fatty acid | TtgR | - | LSR | Metabolic disorders treatment | 63 | |
| Food additive | |||||||
| Off | Phloretin | TtgR | VP16 | TtgA | Biopharmaceutical manufacturing, gene- and cell- based therapies | 8 | |
| On | Vanillic acid | MOR9-1 | - | - | Cell differentiation | 64 | |
| On and Off | Vanillic acid | VanR | VP16 and KRAB | VanA1 and VanA4 | Biopharmaceutical manufacturing, gene- and cell- based therapies | 65 | |
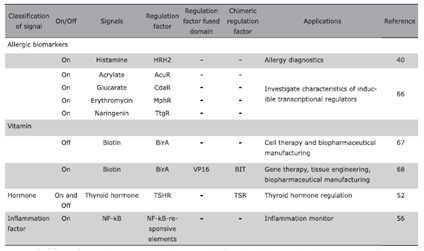
| Allergic biomarkers | |||||||
| On | Histamine | HRH2 | - | - | Allergy diagnostics | 40 | |
| On | Acrylate | AcuR | - | - | Investigate characteristics of inducible transcriptional regulators | 66 | |
| On | Glucarate | CdaR | - | - | |||
| On | Erythromycin | MphR | - | - | |||
| On | Naringenin | TtgR | - | - | |||
| Vitamin | |||||||
| Off | Biotin | BirA | - | - | Cell therapy and biopharmaceutical manufacturing | 67 | |
| On | Biotin | BirA | VP16 | BIT | Gene therapy, tissue engineering, biopharmaceutical manufacturing | 68 | |
| Hormone | On and Off | Thyroid hormone | TSHR | - | TSR | Thyroid hormone regulation | 52 |
| Inflammation factor | On | NF-kB | NF-kB-responsive elements | - | - | Inflammation monitor | 56 |
Continue to Table 1 Characteristics of various simple synthetic biosensors
 |
Note: MphR(A), erythromycin repressor; VP16, Herpes simplex virus virion protein 16; KRAB, Krueppel associated box; ET, erythromycin-dependent transactivator; Pip, pristinamycin-induced protein; PIT, Pristinamycin I-dependent transactivator; ArgR, arginine regulator; ART, arginine-regulated transgene; TrpR, L-tryptophan repressor; TRT,L-tryptophan-dependent transactivator CymR, cumate repressor; cTA, transactivator formed by the fusion of CymR with VP16; rcTA, reverse cumate activator; AlcR, acetaldehyde repressor; HdnoR, 6HNic-responsive repressor; TDAG8,T cell death associated gene 8; EthR, Baeyer-Villiger monooxygenase repressor; ScbR, Streptomyces coelicolor quorum-sensing receptor; SCA, transactivator formed by the fusion of ScbR with VP16; SPA, S.pristinaespiralis-derived transactivator; SpbR, S.pristinaespiralis butyrolactone transcriptional repressor; HucR, bacterial transcriptional repressor that binds a DNA sequence motif (hucO); mUTS, mammalian urate-dependent transsilencer; TGR5(GPBAR1), G protein-coupled bile acid receptor 1; LSR, intracellular lipid-sensing receptor; TtgR, naringenin repressor; TtgA, activator binds and activates transcription from chimeric promoter PTtgR1 harboring OTtgR; MOR9-1, vanillic acid-sensitive olfactory G protein-coupled receptor; VanR, vanillic acid-responsive transcriptional repressor; VanA, activator consisting of VanR fused to VP16; HRH2, the histamine receptor H2; AcuR, acrylate repressor; CdaR, glucarate repressor; MphR, erythromycin repressor; BirA, biotin ligase; BIT, biotin-dependent transactivator; TSHR, thyroid-stimulating hormone receptor; TSR, thyroid-sensing receptor; NF-kB, nuclear factor-k-gene binding.
COMPLEX SYNTHETIC BIOSENSORS
All of the simple synthetic biosensors mentioned above are minimal control devices that can only perform simple tasks involving one signal input. These simple control devices can be assembled into higher order gene circuits which can respond to multiple signal inputs and perform more sophisticated tasks. Multi-signal inputs with complex trigger controlled topology have been designed, including cascade biosensors, Boolean gate biosensors,22 time-delay biosensors,23 hysteretic biosensors and oscillator biosensors.24-26
Cascade biosensors
A cascade synthetic biosensor consists of an artificial stepwise signal cascade whereby the transcription factor at the upper level controls expression of the transcription factor at the lower level that modulates transcription of the target gene at the lowest level.27Each transcription factor at different levels can be induced in an active or a repressive manner by specific signals, thus providing discrete multilevel control in response to different input signals. Fig. 2A illustrates a three-level cascade synthetic biosensor. At the first level, the TET-responsive promoter PTET drives expression of the TET-triggered transactivator tTA and erythromycin (EM)-triggered transactivator, ET1. At the second level, ET1 actives the EM-responsive promoter PERT which controls transcription of the streptogramin-triggered transactivator, a pristinamycin I (PI)-VP16 fusion protein (PIT). At the third level, PIT actives the expression of the reporter, human placental secreted alkaline phosphatase (SEAP). This signaling cascade synthetic biosensor can be interrupted by TET, EM, or PI. They specially inactivate the transcription factor at each level, which are tTA, ET1, and PIT, respectively. The cascade bioreactor is the most basic complex biosensor. It is utilized to process entirely external signals to create the desired function in host by integrating a series of artificial synthetic networks to endogenous physiologic signals.
Boolean gate biosensors
Boolean gate biosensors consist of different compatible ON-type or OFF-type control devices interacting in a parallel or serial combination manner. It can respond to two different input signals (input A, input B) and the relationship of its inputs and outputs can be described using the Boolean operator truth table.28,29 To date, almost all kinds of electronic Boolean gates topology can be realized in synthetic gene networks, including (i) AND gates which are activated when both input A and input B are present,30 (ii) OR gates which are induced when either input A or input B are present, (iii) NOR gates which are repressed when either input A or input B are present, (iv) NOTIF gates which are exclusively induced if input A is present but NOT input B and (v) NAND gates which are always on unless input A and input B are present (Fig. 2B). These basic Boolean gate biosensors can be further assembled to perform more complex tasks by varying the interconnectivity.31
Time-delay biosensors
In time-delayed biosensors, expression of the reporter can be controlled in a time delayed manner. When applying a small amount of signals to the system, persistence of the reporter can be observed even after removal of the signal. Time-delay biosensors are utilized to modulate essential natural biologic patterns, such as control of rhythmic gene expression,32 operation of the circadian clock,33 spacio-temporal coordination of cell differentiation and development, etc. Fig. 2C illustrates a time-delay biosensor.23 The output signal is controlled by biotin in a time-delay manner. The TetR and VP16 domains are respectively fused to streptavidin (SA) and AviTag peptide (AT), forming two new transcriptional regulators: TetR-SA and AT-VP16. In the presence of the Escherichia coli biotin ligase BirA, biotin is able to bind to SA and AT, thereby reconstituting a functional heterodimerize transcription factor, TetR-SA-biotin-AT-VP16, activating the expression of the output signal SEAP. Since the heterodimerization is reversible, the transcription factor TetR-SA-biotin-AT-VP16 can retain its activation function after removal of the signal, biotin, thereby the expression of the downstream reporter SEAP exhibits a time-delay manner.
Oscillator biosensors
Oscillation is of prime importance in biology. In mammals, several processes including endocrine production and release, body temperature modulation, and immune responses that show circadian oscillatory behavior.34,35 The aim of oscillator biosensor is to achieve self-sustaining oscillation of reporter(s) between two separate states. Oscillator biosensors can dynamically control the reporter expression in a periodic pulsed manner without triggering the signal via utilization of different transcriptional activators and repressors. Fig. 2D illustrates a mammalian oscillator biosensor regulated by the tetracycline-dependent transactivator (tTA). It consists of three independent transcription modules: (i) a tTA controlled by a dual promotor system. In this module, the expression of tTA is triggered by the tetracycline-responsive promoter (PhCMV*-1) and inhibited by the streptogramin-responsive promoter (PPIR), (ii) a PhCMV*-1-driven PIT (streptogramin-dependent transactivator) and (iii) a PhCMV*-1-driven destabilized green fluorescent protein (dGFP). The tTA regulation module and the generate a autoregulated oscillating expression loop, therefore the dGFP level whose expression is triggered by tTA shows an oscillatory fashion.24
Hysteretic biosensors
Classic synthetic biosensors provide a dose-dependent graded expression profile in response to signals. In hysteretic biosensors, the threshold required to switch from one state to another depends on the historical environment. Hysteretic biosensor can be utilized for many important natural processes such as the cell cycle36and cell fate control.37 It exhibits the following two characteristics: (i) Threshold levels for ‘ON’ and ‘OFF’ states depend on the starting state rather than the absolute concentration of the inducer. (ii) Different inducer concentrations are required for the system switching from OFF state to the ON state and from the ON state to the OFF state. Fig. 2E illustrates a typical hysteretic biosensor. It consists of a transactivator TetR-VP16 inducing the expression of reporter SEAP and a transsilencer E-KRAB inhibiting the expression of reporter SEAP. TetR-VP16 and E-KRAB compete for binding to the same hybrid promoter PHYBRID which drives the expression of the reporter SEAP and the transactivator TetR-VP16. E-KRAB is constitutively expressed, represses promoter PHYBRID in an erythromycin (EM)-triggered manner. The expression pattern of reporter SEAP depends on the starting state of EM concentrations. High historical EM levels result in high TetR-VP16 levels that require greater E-KRAB activity and lower EM concentrations for an ON-to-OFF switch. Conversely, low historical EM concentrations result in minimal TetR-VP16 levels that require maximum inactivation E-KRAB, hence relatively much higher EM concentrations, for an OFF-to-ON switch.
A. Cascade biosensor. The expression of the output reporter SEAP can be fine-tuned by a three level transcriptional gene circuit and can be interrupted by TET, EM, or PIP. pA, poly(A) tail; IRES, internal ribosome entry site. B. Boolean gate biosensors. PA+Brepresents a promoter that can be triggered when both A and B are present. PA/B represents a promoter that can be triggered by A or by B. C. Time-delay biosensor. The expression of downstream reporter SEAP shows a time-delay manner after removal of biotin from the culture environment. D. Oscillator biosensor. The reporter dGFP level shows an oscillatory manner. E. Hysteretic biosensor. The expression of reporter SEAP exhibits a bistable hysteretic manner. Inducer EM required to switch the reporter from OFF state to ON state is higher than that required for ON to OFF switch.
APPLICATION OF BIOSENSORS IN DISEASE DETECTION
Application of synthetic biosensor as an alternative therapy to conventional pharmacotherapy is promising.38 The basic principle is to utilize the biosensor as a sensor-actuator device which could monitor disease signals in vivo and make therapeutic responses automatically.39 Here, we highlighted some synthetic bio-logy strategies that have been developed to target allergy, liver injuries, insulin resistance, Graves’ disease, urate homeostasis and inflammation.
Histamine-specific allergy
Histamine is the biochemical that is released as part of an allergic reaction, causing swelling, rashes and itching, sneezing, sickness, diarrhea, stomach pains and respiratory problems. Histamine mediates allergic symptoms by binding and activating a family of G protein-coupled receptors, histamine receptors.40 Ausländer et al. constructed a synthetic allergy biosensor by rewiring histamine input to the production of reporter protein, thereby integrating histamine levels with remarkable sensitivity and a wide dynamic range.40 This biosensor consists of a synthetic histamine-responsive signaling cascade in which the G protein-coupled receptor senses extracellular histamine levels and triggers G protein-mediated activation of adenylate cyclase, which in turn converts ATP to cyclic AMP (cAMP). This second messenger molecule binds regulatory subunits of protein kinase A, whose catalytic subunits translocate into the nucleus where they phosphorylate the cAMP-responsive binding protein, which binds and activates synthetic promoter PCREdriving reporter gene expression.
Liver injury
Liver injuries impair the clearance of bile acids from the hepatic portal vein which leads to activation of the G protein-coupled bile acid receptor TGR5 and therefore initiation of a variety of hepatoprotective processes.41-43 Peng et al. created a closed-loop biosensor which could sense the excessive bile acid levels associated with liver injuries and automatically produce a therapeutic protein in response44. They linked activation of ectopically expressed TGR5 to an artificial promoter, PCRE, which controls transcription of the hepatocyte growth factor in a self-sufficient, reversible and dose-dependent manner.
Insulin resistance
Obesity-induced insulin resistance is the key etiologic defect of the metabolic syndrome, a cluster of clinical findings including hypertension and dyslipidaemia, increasing the imminence of cardiovascular disorders and type 2 diabetes.45,46 Ye et al. designed an insulin biosensor aiming to combat the insulin resistance.47 This biosensor consists of an insulin receptor which senses the insulin level and a reporter gene driven by transcription factor TetR-ELK1.48,49 It was designed by rewiring the insulin level to the native IRS-1-Ras-MAPK pathway, activation of which phosphorylates TetR-ELK1, thus triggers transcription from synthetic promoters containing multiple TetR-ELK1-binding sites.49,50
Graves’ disease
Graves’ disease is caused by autoantibodies that activate the thyroid-stimulating hormone (TSH) receptor and trigger a chronic increase in thyroid hormone levels.51 Saxena et al. designed a biosensor to sense thyroid hormone levels and produce an engineered variant of the human TSH, thereby restore the thyrotrophic feedback control of the hypothalamus-pituitary-thyroid axis.52 This biosensor consists of a hormone-sensing receptor (TSR) which precisely monitors thyroid hormone levels and a responding module which regulates the activation of the thyroid hormone receptor. In the absence of the thyroid hormones, TSR is supposed to bind to its cognate promoter and repress gene expression by recruiting endogenous corepressors so that to trigger histone deacetylation and inhibit gene repression. In the presence of thyroid, TSR interacted with coactivators that trigger histone acetylation and mediate gene expression.
Urate homeostasis
Urate is the end-product of purine metabolism. Elevated uric acid levels are associated with gouty arthritis.53 Kemmer et al. engineered a synthetic biosensor to restore uric acid homeostasis in the blood by sensing and responding to uric acid level.54 The detection module of the biosensor is a bacterial transcriptional repressor HucR that binds a DNA sequence motif hucO in the absence of uric acid. The signaling module is a secretion-engineered Aspergillus flavus urate oxidase regulated by hucO. When uric acid is present, HucR dissociates from DNA, thereby allowing expression of a downstream gene, urate oxidase that converts uric acid to allantoin, thereby restoring uric acid homeostasis in the blood.
Inflammation
The design principle of the inflammation biosensor is converting the presence of proinflammatory cytokines to anti-inflammatory cytokines. Schukur et al. designed a biosensor which could sense the presence of tumor necrosis factor (TNF) and interleukin 22 (IL22) levels and produce the anti-inflammatory cytokines IL4 and IL10 with AND-gate-expression logic. This biosensor is applied as an antipsoriatic cytokine converter.55 Based on the same principle, Smole et al. also created an inflammation biosensor comprising a sensor, an amplifier and an effector. This biosensor could be autonomously activated by inflammatory signals and reset externally by a chemical signal.56
FUTURE PERSPECTIVE AND CHANLLENGES
Synthetic biology has showed great potential in disease detection due to its compatibility, orthogonality and fine-tuning ability. Biomedical and clinical applications will necessitate more complex synthetic circuits and construct to perform sophisticated tasks. Although a variety of regulation systems have been identified, most of them are from microbes. Signals associated with clinical diseases are still limited. Therefore, more efforts should be made in identifying and constructing clinically relevant regulation systems. Moreover, as the complexity of a synthetic biosensor increases, computational tools need to be developed to aid the design process in order to guarantee the precision of the synthetic biosensor. The ultimate goal of a sophisticated synthetic biosensor is to play as a therapeutic sensor-effector device that connects diagnostic input with therapeutic output and therefore provides all-in-one diagnostic and therapeutic solutions for future gene- and cell-based therapies.
Conflict of interests statement
All authors declared no conflict of interests.
参考文献
Synthetic biology devices for in vitro and in vivo diagnostics
There is a growing need to enhance our capabilities in medical and environmental diagnostics. Synthetic biologists have begun to focus their biomolecular engineering approaches toward this goal, offering promising results that could lead to the development of new classes of inexpensive, rapidly deployable diagnostics. Many conventional diagnostics rely on antibody-based platforms that, although exquisitely sensitive, are slow and costly to generate and cannot readily confront rapidly emerging pathogens or be applied to orphan diseases. Synthetic biology, with its rational and short design-to-production cycles, has the potential to overcome many of these limitations. Synthetic biology devices, such as engineered gene circuits, bring new capabilities to molecular diagnostics, expanding the molecular detection palette, creating dynamic sensors, and untethering reactions from laboratory equipment. The field is also beginning to move toward in vivo diagnostics, which could provide near real-time surveillance of multiple pathological conditions. Here, we describe current efforts in synthetic biology, focusing on the translation of promising technologies into pragmatic diagnostic tools and platforms.
Genetic regulatory mechanisms in the synthesis of proteins
The synthesis of enzymes in bacteria follows a double genetic control. The socalled structural genes determine the molecular organization of the proteins. Other, functionally specialized, genetic determinants, called regulator and operator genes, control the rate of protein synthesis through the intermediacy of cytoplasmic components or repressors. The repressors can be either inactivated (induction) or activated (repression) by certain specific metabolites. This system of regulation appears to operate directly at the level of the synthesis by the gene of a shortlived intermediate, or messenger, which becomes associated with the ribosomes where protein synthesis takes place.
The TetR family of transcriptional repressors
We have developed a general profile for the proteins of the TetR family of repressors. The stretch that best defines the profile of this family is made up of 47 amino acid residues that correspond to the helix-turn-helix DNA binding motif and adjacent regions in the three-dimensional structures of TetR, QacR, CprB, and EthR, four family members for which the function and three-dimensional structure are known. We have detected a set of 2,353 nonredundant proteins belonging to this family by screening genome and protein databases with the TetR profile. Proteins of the TetR family have been found in 115 genera of gram-positive, alpha-, beta-, and gamma-proteobacteria, cyanobacteria, and archaea. The set of genes they regulate is known for 85 out of the 2,353 members of the family. These proteins are involved in the transcriptional control of multidrug efflux pumps, pathways for the biosynthesis of antibiotics, response to osmotic stress and toxic chemicals, control of catabolic pathways, differentiation processes, and pathogenicity. The regulatory network in which the family member is involved can be simple, as in TetR (i.e., TetR bound to the target operator represses tetA transcription and is released in the presence of tetracycline), or more complex, involving a series of regulatory cascades in which either the expression of the TetR family member is modulated by another regulator or the TetR family member triggers a cell response to react to environmental insults. Based on what has been learned from the cocrystals of TetR and QacR with their target operators and from their three-dimensional structures in the absence and in the presence of ligands, and based on multialignment analyses of the conserved stretch of 47 amino acids in the 2,353 TetR family members, two groups of residues have been identified. One group includes highly conserved positions involved in the proper orientation of the helix-turn-helix motif and hence seems to play a structural role. The other set of less conserved residues are involved in establishing contacts with the phosphate backbone and target bases in the operator. Information related to the TetR family of regulators has been updated in a database that can be accessed at www.bactregulators.org.
Tight control of gene expression in mammalian cells by tetracycline-responsive promoters
Control elements of the tetracycline-resistance operon encoded in Tn10 of Escherichia coli have been utilized to establish a highly efficient regulatory system in mammalian cells. By fusing the tet repressor with the activating domain of virion protein 16 of herpes simplex virus, a tetracycline-controlled transactivator (tTA) was generated that is constitutively expressed in HeLa cells. This transactivator stimulates transcription from a minimal promoter sequence derived from the human cytomegalovirus promoter IE combined with tet operator sequences. Upon integration of a luciferase gene controlled by a tTA-dependent promoter into a tTA-producing HeLa cell line, high levels of luciferase expression were monitored. These activities are sensitive to tetracycline. Depending on the concentration of the antibiotic in the culture medium (0-1 g/ml), the luciferase activity can be regulated over up to five orders of magnitude. Thus, the system not only allows differential control of the activity of an individual gene in mammalian cells but also is suitable for creation of "on/off" situations for such genes in a reversible way.
Daoud-EI Baba M, et al. Macrolide- based transgene control in mammalian cells and mice
Heterologous mammalian gene regulation systems for adjustable expression of multiple transgenes are necessary for advanced human gene therapy and tissue engineering, and for sophisticated in vivo gene-function analyses, drug discovery, and biopharmaceutical manufacturing. The antibiotic-dependent interaction between the repressor (E) and operator (ETR) derived from an Escherichia coli erythromycin-resistance regulon was used to design repressible (E(OFF)) and inducible (E(ON)) mammalian gene regulation systems (E.REX) responsive to clinically licensed macrolide antibiotics (erythromycin, clarithromycin, and roxithromycin). The E(OFF) system consists of a chimeric erythromycin-dependent transactivator (ET), constructed by fusing the prokaryotic repressor E to a eukaryotic transactivation domain that binds and activates transcription from ETR-containing synthetic eukaryotic promoters (P(ETR)). Addition of macrolide antibiotic results in repression of transgene expression. The E(ON) system is based on E binding to artificial ETR-derived operators cloned adjacent to constitutive promoters, resulting in repression of transgene expression. In the presence of macrolides, gene expression is induced. Control of transgene expression in primary cells, cell lines, and microencapsulated human cells transplanted into mice was demonstrated using the E.REX (E(OFF) and E(ON)) systems. The macrolide-responsive E.REX technology was functionally compatible with the streptogramin (PIP-regulated and tetracycline (TET-regulated expression systems, and therefore may be combined for multiregulated multigene therapeutic interventions in mammalian cells and tissues.
Streptogramin-based gene regulation systems for mammalian cells
Here we describe repressible (PipOFF) as well as inducible (PipON) systems for regulated gene expression in mammalian cells, based on the repressor Pip (pristinamycin-induced protein), which is encoded by the streptogramin resistance operon of . Expression of genes placed under control of these systems was responsive to clinically approved antibiotics belonging to the streptogramin group (pristinamycin, virginiamycin, and Synercid). The versatility of these systems was demonstrated by streptogramin-regulated expression of erythropoietin (), placental secreted (SEAP), or () in diverse cell lines (BHK, , HeLa, and myoblasts). Analysis of isogenic constructs in cells demonstrated the PipOFF system gave lower background and higher induction ratios than the widely used -repressible (TetOFF) expression systems. The streptogramin-based expression technology was functionally compatible with the TetOFF system, thus enabling the selective use of different antibiotics to independently control two different gene activities in the same cell.
An engineered L-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice
For optimal compatibility with biopharmaceutical manufacturing and gene therapy, heterologous transgene control systems must be responsive to side-effect-free physiologic inducer molecules. The arginine-inducible interaction of the ArgR repressor and the ArgR-specific ARG box, which synchronize arginine import and synthesis in the intracellular human pathogenChlamydia pneumoniae, was engineered for arginine-regulated transgene (ART) expression in mammalian cells. A synthetic arginine-responsive transactivator (ARG), consisting of ArgR fused to theHerpes simplexVP16 transactivation domain, reversibly adjusted transgene transcription of chimeric ARG box-containing mammalian minimal promoters (PART) in an arginine-inducible manner. Arginine-controlled transgene expression showed rapid induction kinetics in a variety of mammalian cell lines and was adjustable and reversible at concentrations which were compatible with host cell physiology. ART variants containing different transactivation domains, variable spacing between ARG box and minimal promoter and several tandem ARG boxes showed modified regulation performance tailored for specific expression scenarios and cell types. Mice implanted with microencapsulated cells engineered for ART-inducible expression of the human placental secreted alkaline phosphatase (SEAP) exhibited adjustable serum phosphatase levels after treatment with different arginine doses. Using a physiologic inducer, such as the amino acidl-arginine, to control heterologous transgenes in a seamless manner which is devoid of noticeable metabolic interference will foster novel opportunities for precise expression dosing in future gene therapy scenarios as well as the manufacturing of difficult-to-produce protein pharmaceuticals.
Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin
Adjustable control of therapeutic transgenes in engineered cell implants after transdermal and topical delivery of nontoxic trigger molecules would increase convenience, patient compliance, and elimination of hepatic first-pass effect in future therapies. Pseudomonas putida DOT-T1E has evolved the flavonoid-triggered TtgR operon, which controls expression of a multisubstrate-specific efflux pump (TtgABC) to resist plant-derived defense metabolites in its rhizosphere habitat. Taking advantage of the TtgR operon, we have engineered a hybrid P. putida ammalian genetic unit responsive to phloretin. This flavonoid is contained in apples, and, as such, or as dietary supplement, regularly consumed by humans. The engineered mammalian phloretin-adjustable control element (PEACE) enabled adjustable and reversible transgene expression in different mammalian cell lines and primary cells. Due to the short half-life of phloretin in culture, PEACE could also be used to program expression of difficult-to-produce protein therapeutics during standard bioreactor operation. When formulated in skin lotions and applied to the skin of mice harboring transgenic cell implants, phloretin was able to fine-tune target genes and adjust heterologous protein levels in the bloodstream of treated mice. PEACE-controlled target gene expression could foster advances in biopharmaceutical manufacturing as well as gene- and cell-based therapies.
Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice
Prokaryotic transcriptional regulatory elements have been adopted for controlled expression of cloned genes in mammalian cells and animals, the cornerstone for gene-function correlations, drug discovery, biopharmaceutical manufacturing as well as advanced gene therapy and tissue engineering. Many prokaryotes have evolved specific molecular communication systems known as quorum-sensing to coordinate population-wide responses to physiological and/or physicochemical signals. A generic bacterial quorum-sensing system is based on a diffusible signal molecule that prevents binding of a repressor to corresponding operator sites thus resulting in derepression of a target regulon. In Streptomyces, a family of butyrolactones and their corresponding receptor proteins, serve as quorum-sensing systems that control morphological development and antibiotic biosynthesis. Fusion of the Streptomyces coelicolor quorum-sensing receptor (ScbR) to a eukaryotic transactivation domain (VP16) created a mammalian transactivator (SCA) which binds and adjusts transcription from chimeric promoters containing an SCA-specific operator module (P(SPA)). Expression of erythropoietin or the human secreted alkaline phosphatase (SEAP) by this quorum-sensor-regulated gene expression system (QuoRex) could be fine-tuned by non-toxic butyrolactones in a variety of mammalian cells including human primary and mouse embryonic stem cells. Following intraperitoneal implantation of microencapsulated Chinese hamster ovary cells transgenic for QuoRex-controlled SEAP expression into mice, the serum levels of this model glycoprotein could be adjusted to desired concentrations using different butyrolactone dosing regimes.
A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR
Modular design of artificial tissue homeostasis: robust control through synthetic cellular heterogeneity
Abstract Synthetic biology efforts have largely focused on small engineered gene networks, yet understanding how to integrate multiple synthetic modules and interface them with endogenous pathways remains a challenge. Here we present the design, system integration, and analysis of several large scale synthetic gene circuits for artificial tissue homeostasis. Diabetes therapy represents a possible application for engineered homeostasis, where genetically programmed stem cells maintain a steady population of -cells despite continuous turnover. We develop a new iterative process that incorporates modular design principles with hierarchical performance optimization targeted for environments with uncertainty and incomplete information. We employ theoretical analysis and computational simulations of multicellular reaction/diffusion models to design and understand system behavior, and find that certain features often associated with robustness (e.g., multicellular synchronization and noise attenuation) are actually detrimental for tissue homeostasis. We overcome these problems by engineering a new class of genetic modules for 'synthetic cellular heterogeneity' that function to generate beneficial population diversity. We design two such modules (an asynchronous genetic oscillator and a signaling throttle mechanism), demonstrate their capacity for enhancing robust control, and provide guidance for experimental implementation with various computational techniques. We found that designing modules for synthetic heterogeneity can be complex, and in general requires a framework for non-linear and multifactorial analysis. Consequently, we adapt a 'phenotypic sensitivity analysis' method to determine how functional module behaviors combine to achieve optimal system performance. We ultimately combine this analysis with Bayesian network inference to extract critical, causal relationships between a module's biochemical rate-constants, its high level functional behavior in isolation, and its impact on overall system performance once integrated.
The TetR family of regulators
Regulatable systems: applications in gene therapy and replicating viruses
J Clin Invest. 2000 May;105(9):1177-83. Review
Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity
Regulatory elements that control tetracycline resistance in Escherichia coli were previously converted into highly specific transcription regulation systems that function in a wide variety of eukaryotic cells. One tetracycline repressor (TetR) mutant gave rise to rtTA, a tetracycline-controlled transactivator that requires doxycycline (Dox) for binding to tet operators and thus for the activation of Ptetpromoters. Despite the intriguing properties of rtTA, its use was limited, particularly in transgenic animals, because of its relatively inefficient inducibility by doxycycline in some organs, its instability, and its residual affinity to tetO in absence of Dox, leading to elevated background activities of the target promoter. To remove these limitations, we have mutagenized tTA DNA and selected in Saccharomyces cerevisiae for rtTA mutants with reduced basal activity and increased Dox sensitivity. Five new rtTAs were identified, of which two have greatly improved properties. The most promising new transactivator, rtTA2s-M2, functions at a 10-fold lower Dox concentration than rtTA, is more stable in eukaryotic cells, and causes no background expression in the absence of Dox. The coding sequences of the new reverse TetR mutants fused to minimal activation domains were optimized for expression in human cells and synthesized. The resulting transactivators allow stringent regulation of target genes over a range of 4 to 5 orders of magnitude in stably transfected HeLa cells. These rtTA versions combine tightness of expression control with a broad regulatory range, as previously shown for the widely applied tTA.
Pluripotentiality and conditional transgene regulation in human embryonic stem cells expressing insulated tetracycline-ON transactivator
Disclosure of potential conflicts of interest is found at the end of this article.
Generation of pig induced pluripotent stem cells with a drug-inducible system
【CateGory Index】: R329
Establishment of tetracycline-inducible, survivin-expressing CHO cell lines by an optimized screening method
An optimized method based on tetracycline-inducible gene expression system T-REx was developed to screen and evaluate Tet repressor (TetR)-expressing cell lines using enhanced green fluorescence protein (EGFP) as reporter gene. To verify the effectiveness of the method, two TetR-expressing Chinese hamster ovary (CHO) cell lines, CHO-TR B2 (stringent) and B5 (less stringent), in which the EGFP genes were variantly controlled by tetracycline, were used to construct cell lines expressing the anti-apoptosis gene survivin upon induction with tetracycline. The resulting stable clones were analyzed for survivin expression. The analysis showed that all four B5-derived clones exhibited leaky survivin expression in the absence of tetracycline, while the B2-derived clones did not. DNA laddering and annexin V/PI staining assays further indicated that although tetracycline-inducible expression of survivin conferred resistance to NHl- and staurosporine-induced apoptosis in both the B2- and the B5-derived stable cell lines, the B2-derived cell lines showed more stringent regulation in the absence of tetracycline. This represents successful utilization of the present screening method.
Optimization of tetracycline-responsive recombinant protein production and effect on cell growth and ER stress in mammalian cells
The inducible T-REx system and other inducible expression systems have been developed in order to control the expression levels of recombinant protein in mammalian cells. In order to study the effects of heterologous protein expression on mammalian host behavior, the gene for recombinant Human transferrin (hTf) was integrated into HEK-293 cells and expressed under the control of the T-REx inducible technology (293-TetR-Hyg-hTf) or using a constitutive promoter (293-CMV-hTf). A number of inducible clones with variable expression levels were identified for the T-REx system with levels of hTf for the high expressing clones nearly double those obtained using the constitutive cytomegalovirus (CMV) promoter. The level of transferrin produced was found to increase proportionately with tetracycline concentration between 0 and 1 g/mL with no significant increases in transferrin production above 1 g/mL. As a result, the optimal induction time and tetracycline concentrations were determined to be the day of plating and 1 g/mL, respectively. Interestingly, the cells induced to express transferrin, 293-TetR-Hyg-hTf, exhibited lower viable cell densities and percent viabilities than the uninduced cultures for multiple clonal isolates. In addition, the induction of transferrin expression was found to cause an increase in the expression of the ER-stress gene, BiP , that was not observed in the uninduced cells. However, both uninduced and induced cell lines containing the hTf gene exhibited longer survival in culture than the control cells, possibly as a result of the positive effects of hTf on cell survival. Taken together, these results suggest that the high level expression of complex proteins in mammalian cells can limit the viable cell densities of cells in culture as a result of cellular stresses caused by generating proteins that may be difficult to fold or are otherwise toxic to cells. The application of inducible systems such as the T-REx technology will allow us to optimize protein production while limiting the negative effects that result from these cellular stresses. 2005 Wiley Periodicals, Inc.
Tetracycline-induci-ble expression systems with reduced basal activity in mammalian cells
We describe a modification of the tetracycline-inducible eukaryotic gene expression system with decreased basal levels of expression in HeLa cells. It employs the tetracycline-inducible transactivator and a tetracycline-regulated repressor fusion acting on the same promoter. To avoid heterodimerization or competition for the same DNA site, each was provided with different DNA recognition and/or protein dimerization specificities. We achieved active silencing in the uninduced state resulting in approximately 6-fold reduced levels of basal transcription and several hundred-fold activation of gene expression upon addition of tetracycline.
A more efficient RNAi inducible system for tight regulation of gene expression in mammalian cells and xenograft animals
Abstract Two types of tetracycline-controlled inducible RNAi expression systems have been developed that generally utilize multiple tetracycline operators (TetOs) or repressor fusion proteins to overcome the siRNA leakiness. Here, we report a novel system that overexpresses the tetracycline repressor (TetR) via a bicistronic construct to control siRNA expression. The high level of TetR expression ensures that the inducible promoter is tightly bound, with minimal basal transcription, allowing for regulation solely dependent on TetR rather than a TetR fusion protein via a more complicated mechanism. At the same time, this system contains only a single TetO, thus minimizing the promoter impairment occurring in existing systems due to the incorporation of multiple TetOs, and maximizing the siRNA expression upon induction. In addition, this system combines all the components required for regulation of siRNA expression into a single lentiviral vector, so that stable cell lines can be generated by a single transduction and selection, with significant reduction in time and cost. Taken together, this all-in-one lentiviral vector with the feature of TetR overexpression provides a unique and more efficient tool for conditional gene knockdown that has wide applications. We have demonstrated the high degree of robustness and versatility of this system as applied to several mammalian cells and xenograft animals.
Recent advances in mammalian synthetic biology-design of synthetic transgene control networks
Capitalizing on an era of functional genomic research, systems biology offers a systematic quantitative analysis of existing biological systems thereby providing the molecular inventory of biological parts that are currently being used for rational synthesis and engineering of complex biological systems with novel and potentially useful functionsn emerging discipline known as synthetic biology. During the past decade synthetic biology has rapidly developed from simple control devices fine-tuning the activity of single genes and proteins to multi-gene/protein-based transcription and signaling networks providing new insight into global control and molecular reaction dynamics, thereby enabling the design of novel drug-synthesis pathways as well as genetic devices with unmatched biological functions. While pioneering synthetic devices have first been designed as test, toy, and teaser systems for use in prokaryotes and lower eukaryotes, first examples of a systematic assembly of synthetic gene networks in mammalian cells has sketched the full potential of synthetic biology: foster novel therapeutic opportunities in gene and cell-based therapies. Here we provide a concise overview on the latest advances in mammalian synthetic biology.
McWhite C, et al. Sensitive detection of proteasomal activation using the Deg-On mammalian synthetic gene circuit
Abstract The ubiquitin proteasome system (UPS) has emerged as a drug target for diverse diseases characterized by altered proteostasis, but pharmacological agents that enhance UPS activity have been challenging to establish. Here we report the Deg-On system, a genetic inverter that translates proteasomal degradation of the transcriptional regulator TetR into a fluorescent signal, thereby linking UPS activity to an easily detectable output, which can be tuned using tetracycline. We demonstrate that this circuit responds to modulation of UPS activity in cell culture arising from the inhibitor MG-132 and activator PA28. Guided by predictive modelling, we enhanced the circuit's signal sensitivity and dynamic range by introducing a feedback loop that enables self-amplification of TetR. By linking UPS activity to a simple and tunable fluorescence output, these genetic inverters will enable a variety of applications, including screening for UPS activating molecules and selecting for mammalian cells with different levels of proteasome activity.
A synthetic time-delay circuit in mammalian cells and mice
Time-delay circuitries in which a transcription factor processes independent input parameters can modulate NF-B activation, manage quorum-sensing cross-talk, and control the circadian clock. We have constructed a synthetic mammalian gene network that processes four different input signals to control either immediate or time-delayed transcription of specific target genes. BirA-mediated ligation of biotin to a biotinylation signal-containing VP16 transactivation domain triggers heterodimerization of chimeric VP16 to a streptavidin-linked tetracycline repressor (TetR). At increasing biotin concentrations up to 20 nM, TetR-specific promoters are gradually activated (off to on, input signal 1), are maximally induced at concentrations between 20 nM and 10/M, and are adjustably shut off at biotin levels exceeding 10/M (on to off, input signal 2). These specific expression characteristics with a discrete biotin concentration window emulate a biotin-triggered bandpass filter. Removal of biotin from the culture environment (input signal 3) results in time-delayed transgene expression until the intracellular biotinylated VP16 pool is degraded. Because the TetR component of the chimeric transactivator retains its tetracycline responsiveness, addition of this antibiotic (input signal 4) overrides biotin control and immediately shuts off target gene expression. Biotin-responsive immediate, bandpass filter, and time-delay transcription characteristics were predicted by a computational model and have been validated in standard cultivation settings or biopharmaceutical manufacturing scenarios using trangenic CHO-K1 cell derivatives and have been confirmed in mice. Synthetic gene circuitries provide insight into structure-function correlations of native signaling networks and foster advances in gene therapy and biopharmaceutical manufacturing.
A tunable synthetic mammalian oscillator
A synthetic low-frequency mammalian oscillator
Circadian clocks have long been known to be essential for the maintenance of physiological and behavioral processes in a variety of organisms ranging from plants to humans. Dysfunctions that subvert gene expression of oscillatory circadian-clock components may result in severe pathologies, including tumors and metabolic disorders. While the underlying molecular mechanisms and dynamics of complex gene behavior are not fully understood, synthetic approaches have provided substantial insight into the operation of complex control circuits, including that of oscillatory networks. Using iterative cycles of mathematical model-guided design and experimental analyses, we have developed a novel low-frequency mammalian oscillator. It incorporates intronically encoded siRNA-based silencing of the tetracycline-dependent transactivator to enable the autonomous and robust expression of a fluorescent transgene with periods of 26 h, a circadian clock-like oscillatory behavior. Using fluorescence-based time-lapse microscopy of engineered CHO-K1 cells, we profiled expression dynamics of a destabilized yellow fluorescent protein variant in single cells and real time. The novel oscillator design may enable further insights into the system dynamics of natural periodic processes as well as into siRNA-mediated transcription silencing. It may foster advances in design, analysis and application of complex synthetic systems in future gene therapy initiatives.<br>
Synthetic memory circuits for tracking human cell fate
Abstract A variety of biological phenomena, from disease progression to stem cell differentiation, are typified by a prolonged cellular response to a transient environmental cue. While biologically relevant, heterogeneity in these long-term responses is difficult to assess at the population level, necessitating the development of biological tools to track cell fate within subpopulations. Here we present a novel synthetic biology approach for identifying and tracking mammalian cell subpopulations. We constructed three genomically integrated circuits that use bistable autoregulatory transcriptional feedback to retain memory of exposure to brief stimuli. These "memory devices" are used to isolate and track the progeny of cells that responded differentially to doxycycline, hypoxia, or DNA-damaging agents. Following hypoxic or ultraviolet radiation exposure, strongly responding cells activate the memory device and exhibit changes in gene expression, growth rates, and viability for multiple generations after the initial stimulus. Taken together, these results indicate that a heritable memory of hypoxia and DNA damage exists in subpopulations that differ in long-term cell behavior.
Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells
Abstract Prototype drug-adjustable heterologous transcription control systems designed for gene therapy applications typically show sigmoid dose–response characteristics and enable fine-tuning of therapeutic transgenes only within a narrow inducer concentration range of a few nanograms. However, the design of clinical dosing regimes which achieve tissue-specific concentrations with nanogram precision is yet a “mission impossible.” Therefore, most of today's transcription control systems operate as ON/OFF switches and not in a true adjustable mode. The availability of robust transcription control configurations which lock expression of a single therapeutic transgene at desired levels in response to fixed clinical doses of different inducers rather than minute concentration changes of a single inducer would be highly desirable. Based on in silico predictions, we have constructed a variety of mammalian artificial regulatory networks by interconnecting the tetracycline- (TET OFF ), streptogramin- (PIP OFF ), and macrolide- (E OFF ) repressible gene regulation systems as linear (auto)regulatory cascades. These networks enable multilevel expression control of several transgenes in response to different antibiotics or allow titration of a single transgene to four discrete expression levels by clinical dosing of a single antibiotic: 1) high expression in the absence of any antibiotic (+++), 2) medium level expression following addition of tetracycline (++), 3) low level expression in response to the macrolide erythromycin (+), and 4) complete repression by streptogramins such as pristinamycin (–). The first-generation artificial regulatory networks exemplify modular interconnections of different heterologous gene regulations systems to achieve multigene expression, fine-tuning, or to design novel control networks with unprecedented transgene regulation properties. Such higher-level transcription control modalities will lead the way towards composite artificial regulatory networks able to effect complex therapeutic interventions in future gene therapy and tissue engineering scenarios. 08 2003 Wiley Periodicals. Biotechnol Bioeng 83: 810–820, 2003.
BioLogic gates enable logical transcription control in mammalian cells
Abstract The architecture of gene regulatory networks is reminiscent of electronic circuits. Modular building blocks that respond in a logical way to one or several inputs are connected to perform a variety of complex tasks. Gene circuit engineers have pioneered the construction of artificial gene regulatory networks with the intention to pave the way for the construction of therapeutic gene circuits for next-generation gene therapy approaches. However, due to the lack of a critical amount of eukaryotic cell-compatible gene regulation systems, the field has so far been limited to prokaryotes. Recent development of several mammalian cell-compatible expression control systems laid the foundations for the assembly of transcription control modules that can respond to several inputs. Herein, three approaches to evoke combinatorial transcription control have been followed: (i) construction of artificial promoters with up to three operator sites for regulatory proteins, and (ii) parallel and (iii) serial linking of two gene regulation systems. We have combined tetracycline-, streptogramin-, macrolide-, and butyrolactone transcription control systems to engineer BioLogic gates of the NOT IF-, AND-, NOT IF IF-, NAND-, OR-, NOR-, and INVERTER-type in mammalian cells, which are able to respond to up to three different small molecule inputs. BioLogic gates enable logical transcriptional control in mammalian cells and, in combination with modern transduction technologies, could serve as versatile tools for regulated gene expression and as building blocks for complex artificial gene regulatory networks for applications in gene therapy, tissue engineering, and biotechnology. 2004 Wiley Periodicals, Inc.
Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation
An engineered commensal E. coli strain can function as a living diagnostic for a marker of inflammation in the murine gut for 200 days.
Synthetic biology platform for sensing and integrating endogenous transcriptional inputs in mammalian cells
One of the goals of synthetic biology is to develop programmable artificial gene networks that can transduce multiple endogenous molecular cues to precisely control cell behavior. Realizing this vision requires interfacing natural molecular inputs with synthetic components that generate functional molecular outputs. Interfacing synthetic circuits with endogenous mammalian transcription factors has been particularly difficult. Here, we describe a systematic approach that enables integration and transduction of multiple mammalian transcription factor inputs by a synthetic network. The approach is facilitated by a proportional amplifier sensor based on synergistic positive autoregulation. The circuits efficiently transduce endogenous transcription factor levels into RNAi, transcriptional transactivation, and site-specific recombination. They also enable AND logic between pairs of arbitrary transcription factors. The results establish a framework for developing synthetic gene networks that interface with cellular processes through transcriptional regulators. Graphical Abstract 61A positive feedback loop with high synergy between trigger input and amplifier61Rule-based design of robust amplified sensors of mammalian transcription factors61AND gates between pairs of unrelated transcription factors61Efficient transduction of transcriptional inputs into diverse downstream actuation A positive feedback loop with high synergy between trigger input and amplifier Rule-based design of robust amplified sensors of mammalian transcription factors AND gates between pairs of unrelated transcription factors Efficient transduction of transcriptional inputs into diverse downstream actuation Coupling endogenous transcription factor activities to synthetic gene circuits has been a longstanding challenge. Angelici et02al. describe highly synergistic composite promoters that enable robust and selective amplification of mammalian transcriptional inputs using positive feedback as well as their arbitrary pairing in promoter-level AND gates. The resulting sensors can efficiently transduce their inputs’ signal to downstream synthetic circuits via a variety of mechanisms, including RNAi, transactivation, and recombination.
Engineering molecular circuits using synthetic biology in mammalian cells
Abstract Synthetic biology has made significant leaps over the past decade, and it now enables rational and predictable reprogramming of cells to conduct complex physiological activities. The bases for cellular reprogramming are mainly genetic control components affecting gene expression. A huge variety of these modules, ranging from engineered fusion proteins regulating transcription to artificial RNA devices affecting translation, is available, and they often feature a highly modular scaffold. First endeavors to combine these modules have led to autoregulated expression systems and genetic cascades. Analogous to the rational engineering of electronic circuits, the existing repertoire of artificial regulatory elements has further enabled the ambitious reprogramming of cells to perform Boolean calculations or to mimic the oscillation of circadian clocks. Cells harboring synthetic gene circuits are not limited to cell culture, as they have been successfully implanted in animals to obtain tailor-made therapeutics that have made it possible to restore urea or glucose homeostasis as well as to offer an innovative approach to artificial insemination.
Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays
Feedback inhibition of gene expression is a widespread phenomenon in which the expression of a gene is downregulated by its protein product. Feedback in eukaryotic cells involves time delays resulting from transcription, transcript splicing and processing, and protein synthesis. In principle, such delays can result in oscillatory mRNA and protein expression [1]. However, experimental evidence of such delay-driven oscillations has been lacking. Using mathematical modeling informed by recent data, I show that the observed oscillatory expression and activity of three proteins is most likely to be driven by transcriptional delays. Each protein (Hes1, p53, and NF-B) is a component of a short feedback inhibition loop . The oscillatory period is determined by the delay and the protein and mRNA half-lives, but it is robust to changes in the rates of transcription and protein synthesis. In contrast to nondelayed models, delayed models do not require additional components in the feedback loop. These results provide direct evidence that transcriptional delays can drive oscillatory gene activity and highlight the importance of considering delays when analyzing genetic regulatory networks, particularly in processes such as developmental pattern formation, where short half-lives and feedback inhibition are common.
Control mechanism of the circadian clock for timing of cell division in vivo
Cell division in many mammalian tissues is associated with specific times of day, but just how the circadian clock controls this timing has not been clear. Here, we show in the regenerating liver (of mice) that the circadian clock controls the expression of cell cycle-related genes that in turn modulate the expression of active Cyclin B1-Cdc2 kinase, a key regulator of mitosis. Among these genes, expression of wee1 was directly regulated by the molecular components of the circadian clockwork. In contrast, the circadian clockwork oscillated independently of the cell cycle in single cells. Thus, the intracellular circadian clockwork can control the cell-division cycle directly and unidirectionally in proliferating cells.
Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity
The pattern of circadian behavioral rhythms is photoperiod-dependent, highlighted by the conservation of a phase relation between the behavioral rhythm and photoperiod. A model of two separate, but mutually coupled, circadian oscillators has been proposed to explain photoperiodic responses of behavioral rhythm in nocturnal rodents: an evening oscillator, which drives the activity onset and entrains to dusk, and a morning oscillator, which drives the end of activity and entrains to dawn. Continuous measurement of circadian rhythms in clock gene Per1 expression by a bioluminescence reporter enabled us to identify the separate oscillating cell groups in the mouse suprachiasmatic nucleus (SCN), which composed circadian oscillations of different phases and responded to photoperiods differentially. The circadian oscillation in the posterior SCN was phase-locked to the end of activity under three photoperiods examined. On the other hand, the oscillation in the anterior SCN was phase-locked to the onset of activity but showed a bimodal pattern under a long photoperiod [light-dark cycle (LD)18:6]. The bimodality in the anterior SCN reflected two circadian oscillatory cell groups of early and late phases. The anterior oscillation was unimodal under intermediate (LD12:12) and short (LD6:18) photoperiods, which was always phase-lagged behind the posterior oscillation when the late phase in LD18:6 was taken. The phase difference was largest in LD18:6 and smallest in LD6:18. These findings indicate that three oscillating cell groups in the SCN constitute regionally specific circadian oscillations, and at least two of them are involved in photoperiodic response of behavioral rhythm.
The daily rhythms of genes, cells and organs
EMBO reports encourages and publishes articles that report novel findings of wide biological significance in the areas of development, immunology, neuroscience, plant biology, structural biology, genomic & computational biology, genome stability & dynamics, chromatin & transcription, RNA, proteins, cellular metabolism, signal transduction, cell cycle, differentiation & death, membranes & transport, cell & tissue architecture, microbiology & pathogens and molecular biology of disease.
Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts
Cells progressing through the cell cycle must commit irreversibly to mitosis without slipping back to interphase before properly segregating their chromosomes. A mathematical model of cell-cycle progression in cell-free egg extracts from frog predicts that irreversible transitions into and out of mitosis are driven by hysteresis in the molecular control system. Hysteresis refers to toggle-like switching behavior in a dynamical system. In the mathematical model, the toggle switch is created by positive feedback in the phosphorylation reactions controlling the activity of Cdc2, a protein kinase bound to its regulatory subunit, cyclin B. To determine whether hysteresis underlies entry into and exit from mitosis in cell-free egg extracts, we tested three predictions of the Novak-Tyson model. (i) The minimal concentration of cyclin B necessary to drive an interphase extract into mitosis is distinctly higher than the minimal concentration necessary to hold a mitotic extract in mitosis, evidence for hysteresis. (ii) Unreplicated DNA elevates the cyclin threshold for Cdc2 activation, indication that checkpoints operate by enlarging the hysteresis loop. (iii) A dramatic "slowing down" in the rate of Cdc2 activation is detected at concentrations of cyclin B marginally above the activation threshold. All three predictions were validated. These observations confirm hysteresis as the driving force for cell-cycle transitions into and out of mitosis.
A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision
Engineering gene circuits for mammalian cell-based applications
Abstract Synthetic gene switches are basic building blocks for the construction of complex gene circuits that transform mammalian cells into useful cell-based machines for next-generation biotechnological and biomedical applications. Ligand-responsive gene switches are cellular sensors that are able to process specific signals to generate gene product responses. Their involvement in complex gene circuits results in sophisticated circuit topologies that are reminiscent of electronics and that are capable of providing engineered cells with the ability to memorize events, oscillate protein production, and perform complex information-processing tasks. Microencapsulated mammalian cells that are engineered with closed-loop gene networks can be implanted into mice to sense disease-related input signals and to process this information to produce a custom, fine-tuned therapeutic response that rebalances animal metabolism. Progress in gene circuit design, in combination with recent breakthroughs in genome engineering, may result in tailored engineered mammalian cells with great potential for future cell-based therapies. Copyright @ 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Applications of synthetic gene networks
Synthetic gene networks have evolved from simple proof-of-concept circuits to complex therapy-oriented networks over the past 15 years. This advancement has greatly facilitated the expansion of the emerging field of synthetic biology. In this review, we highlight the main applications of synthetic gene networks in understanding biological design principles, developing biosensors for diagnosis, producing industrial and biomedical compounds, and treating human diseases. Finally, we outline current challenges and future prospects of synthetic gene networks for advancing practical applications.
A designer cell-based histamine-specific human allergy profiler
Abstract Allergic disorders are markedly increasing in industrialized countries. The identification of compounds that trigger the immunoglobulin E-dependent allergic reaction remains the key to limit patients' exposure to critical allergens and improve their quality of life. Here we use synthetic biology principles to design a mammalian cell-based allergy profiler that scores the allergen-triggered release of histamine from whole-blood-derived human basophils. A synthetic signalling cascade engineered within the allergy profiler rewires histamine input to the production of reporter protein, thereby integrating histamine levels in whole-blood samples with remarkable sensitivity and a wide dynamic range, allowing for rapid results or long-term storage of output, respectively. This approach provides non-intrusive allergy profiles for the personalized medicine era.
Targeting bile-acid signalling for metabolic diseases
Bile acids: trying to understand their chemistry and biology with the hope of helping patients
An informal review of the author's five decades of research on the chemistry and biology of bile acids in health and disease is presented. The review begins with a discussion of bile acid structure and its remarkable diversity in vertebrates. Methods for tagging bile acids with tritium for metabolic or transport studies are summarized. Bile acids solubilize polar lipids in mixed micelles; progress in elucidating the structure of the mixed micelle is discussed. Extensive studies on bile acid metabolism in humans have permitted the development of physiological pharmacokinetic models that can be used to simulate bile acid metabolism. Consequences of defective bile acid biosynthesis and transport have been clarified, and therapy has been developed. Methods for measuring bile acids have been improved. The rise and fall of medical and contact dissolution of cholesterol gallstones is chronicled. Finally, principles of therapy with bile acid agonists and antagonists are given. Advances in understanding bile acid biology and chemistry have helped to improve the lives of patients with hepatobiliary or digestive disease. (H EPATOLOGY 2009.)
The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice
Many regulatory pathways are involved in liver regeneration after partial hepatectomy (PH) to initiate growth, protect liver cells, and sustain functions of the remnant liver. Bile acids (BAs), whose levels rise in the blood early after PH, stimulate both hepatocyte proliferation and protection, in part through their binding to the nuclear farnesoid X receptor (FXR). However, the effect of the BA receptor, TGR5 (G-protein-coupled BA receptor 1) after PH remains to be studied. Liver histology, hepatocyte proliferation, BA concentrations (plasma, bile, liver, urine, and feces), bile flow and composition, and cytokine production were studied in wild-type (WT) and TGR5 KO (knockout) mice before and after PH. BA composition (plasma, bile, liver, urine, and feces) was more hydrophobic in TGR5 KO than in WT mice. After PH, severe hepatocyte necrosis, prolonged cholestasis, exacerbated inflammatory response, and delayed regeneration were observed in TGR5 KO mice. Although hepatocyte adaptive response to post-PH BA overload was similar in WT and TGR5 KO mice, kidney and biliary adaptive responses were strongly impaired in TGR5 KO mice. Cholestyramine treatment, as well as Kupffer cell depletion, significantly improved the post-PH TGR5 KO mice phenotype. After bile duct ligation or upon a cholic acidnriched diet, TGR5 KO mice exhibited more severe liver injury than WT as well as impaired BA elimination in urine. Conclusion: TGR5 is crucial for liver protection against BA overload after PH, primarily through the control of bile hydrophobicity and cytokine secretion. In the absence of TGR5, intrahepatic stasis of abnormally hydrophobic bile and excessive inflammation, in association with impaired bile flow adaptation and deficient urinary BA efflux, lead to BA overload-induced liver injury and delayed regeneration. (Hepatology 2013;58:1451~1460)
A synthetic biology-based device prevents liver injury in mice
The liver performs a panoply of complex activities coordinating metabolic, immunologic and detoxification processes. Despite the liver robustness and unique self-regeneration capacity, viral infection, autoimmune disorders, fatty liver disease, alcohol abuse and drug-induced hepatotoxicity contribute to the increasing prevalence of liver failure. Liver injuries impair the clearance of bile acids from the hepatic portal vein which leads to their spill over into the peripheral circulation where they activate the G-protein-coupled bile acid receptor TGR5 to initiate a variety of hepatoprotective processes. By functionally linking activation of ectopically expressed TGR5 to an artificial promoter controlling transcription of the hepatocyte growth factor (HGF), we created a closed-loop synthetic signalling network that coordinated liver injury-associated serum bile acid levels to expression of HGF in a self-sufficient, reversible and dose-dependent manner. After implantation of genetically engineered human cells inside auto-vascularizing, immunoprotective and clinically validated alginate-poly-(L-lysine)-alginate beads into mice, the liver-protection device detected pathologic serum bile acid levels and produced therapeutic HGF levels that protected the animals from acute drug-induced liver failure. Genetically engineered cells containing theranostic gene circuits that dynamically interface with host metabolism may provide novel opportunities for preventive, acute and chronic healthcare. Liver diseases leading to organ failure may go unnoticed as they do not trigger any symptoms or significant discomfort. We have designed a synthetic gene circuit that senses excessive bile acid levels associated with liver injuries and automatically produces a therapeutic protein in response. When integrated into mammalian cells and implanted into mice, the circuit detects the onset of liver injuries and coordinates the production of a protein pharmaceutical which prevents liver damage.
The origins and drivers of insulin resistance
Obesity-induced insulin resistance is the major determinant of metabolic syndrome, which precedes the development of type 2 diabetes mellitus and is thus the driving force behind the emerging diabetes epidemic. The precise causes of insulin resistance are varied, and the relative importance of each is a matter of ongoing research. Here, we offer a Perspective on the heterogeneous etiology of insulin resistance, focusing in particular on the role of inflammation, lipid metabolism, and the gastrointestinal microbiota.
Mechanisms for insulin resis-tance: common threads and missing links
Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat Bio-med
A self-adjusting synthetic gene circuit in implanted mammalian cells senses insulin concentration and reverses the insulin-resistance syndrome in three mouse models by coordinating the expression of the insulin-sensitizing compound adiponectin.
Insulin receptor tyrosine kinase activity and phosphorylation of tyrosines 1162 and 1163 are required for insulin-increased prolactin gene expression
Insulin treatment increased prolactin gene expression in GH4 cells, a rat pituitary tumor cell line, through the endogenous insulin receptor. However, insulin regulation of transfected plasmids required the expression of cotransfected insulin receptor. Prolactin-CAT expression was increased 12-fold in cells transfected with wild type insulin receptor, but insulin did not increase prolactin gene expression when a kinase negative mutant of the ATP binding site (K1030R) was expressed. Thus, receptor kinase activity was required for signaling to gene transcription. Mutation of tyrosine 1158 did not reduce insulin-increased prolactin-CAT expression while individual mutations of tyrosine 1162 and tyrosine 1163 each reduced insulin-increased prolactin-CAT expression by 50% and a triple mutant of tyrosines 1158/1162/1163 was inactive. Thus, mutation of tyrosine 1162 and 1163 was also sufficient to inactivate signaling by the insulin receptor. Insulin-stimulated auto phosphorylation occurred in all mutants in vitro except the ATP binding site mutant. However, the ability of mutant insulin receptors to mediate insulin-increased prolactin-CAT expression correlated with the substrate-specific catalytic activity of the receptors. This suggested that phosphorylation of these tyrosines was important for substrate access to the catalytic domain of the receptor.
Signalling by insulin and IGF receptors: supporting acts and new players
The signalling pathways utilised by insulin receptor (IR) and IGF receptor to transduce their diverse effects on cellular metabolism, growth and survival are well established in broad outline, but many details remain to be elucidated. Tyrosine phosphorylation of IR substrates and Shc initiates signalling via canonical phosphoinositide 3-kinase/Akt and Ras/MAP kinase pathways, which together mediate many of the actions of insulin and IGFs. However, a variety of additional substrates and scaffolds have been described that may play roles in modulating the canonical pathways or in specific biological responses. This review will focus on recent studies that have extended our understanding of insulin/IGF signalling pathways, and the elements that may contribute to specificity.
TetR hybrid transcription factors report cell signaling and are inhibited by doxycycline
We developed a bigenic reporter system composed of a hybrid transcription factor that combines the regulatory and activation domains of either Elk-1 or cyclic AMP-responsive element binding protein (CREB) with the DNA binding, dimerization, and regulatory domains from a synthetic variant of the bacterial Tet repressor (TetR). The novel hybrid transcription factor TetR-Elk-1 was regulated by MAPK ERK kinase 1 (MEK-1) overexpression, and TetR-CREB was regulated by protein kinase A (PKA) overexpression or elevation of cyclic AMP levels. These hybrid transcription factors could be useful reporters of cell signaling pathways because, unlike previous GAL4 hybrid reporters, TetR hybrid transcription factors are inhibited by the administration of doxycycline. We validated this system in cell culture transfection experiments utilizing luciferase assays to monitor reporter gene expression and Western blot analysis to monitor transcription factor expression and phosphorylation levels. This system may be useful in creating temporally restricted windows of response to cell signaling and may be of value in the advancement of methods used to study signal transduction.
Thyroid hormone regulation of metabolism
Thyroid hormone (TH) is required for normal development as well as regulating metabolism in the adult. The thyroid hormone receptor (TR) isoforms, and , are differentially expressed in tissues and have distinct roles in TH signaling. Local activation of thyroxine (T4), to the active form, triiodothyronine (T3), by 5'-deiodinase type 2 (D2) is a key mechanism of TH regulation of metabolism. D2 is expressed in the hypothalamus, white fat, brown adipose tissue (BAT), and skeletal muscle and is required for adaptive thermogenesis. The thyroid gland is regulated by thyrotropin releasing hormone (TRH) and thyroid stimulating hormone (TSH). In addition to TRH/TSH regulation by TH feedback, there is central modulation by nutritional signals, such as leptin, as well as peptides regulating appetite. The nutrient status of the cell provides feedback on TH signaling pathways through epigentic modification of histones. Integration of TH signaling with the adrenergic nervous system occurs peripherally, in liver, white fat, and BAT, but also centrally, in the hypothalamus. TR regulates cholesterol and carbohydrate metabolism through direct actions on gene expression as well as cross-talk with other nuclear receptors, including peroxisome proliferator-activated receptor (PPAR), liver X receptor (LXR), and bile acid signaling pathways. TH modulates hepatic insulin sensitivity, especially important for the suppression of hepatic gluconeogenesis. The role of TH in regulating metabolic pathways has led to several new therapeutic targets for metabolic disorders. Understanding the mechanisms and interactions of the various TH signaling pathways in metabolism will improve our likelihood of identifying effective and selective targets.
Charpin-El Hamri G, et al. Synthetic gene network restoring endogenous pituitary-thyroid feedback control in experimental Graves’ disease
Daoud-El Baba M, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit
Abstract Synthetic biology has shown that the metabolic behavior of mammalian cells can be altered by genetic devices such as epigenetic and hysteretic switches, timers and oscillators, biocomputers, hormone systems and heterologous metabolic shunts. To explore the potential of such devices for therapeutic strategies, we designed a synthetic mammalian circuit to maintain uric acid homeostasis in the bloodstream, disturbance of which is associated with tumor lysis syndrome and gout. This synthetic device consists of a modified Deinococcus radiodurans-derived protein that senses uric acids levels and triggers dose-dependent derepression of a secretion-engineered Aspergillus flavus urate oxidase that eliminates uric acid. In urate oxidase-deficient mice, which develop acute hyperuricemia, the synthetic circuit decreased blood urate concentration to stable sub-pathologic levels in a dose-dependent manner and reduced uric acid crystal deposits in the kidney. Synthetic gene-network devices providing self-sufficient control of pathologic metabolites represent molecular prostheses, which may foster advances in future gene- and cell-based therapies.
7(318):318ra201
doi:
A Synthetic Mammalian Therapeutic Gene Circuit for Sensing and Suppressing Inflammation
Inflammation, which is a highly regulated host response against danger signals, may be harmful if it is excessive and deregulated. Ideally, anti-inflammatory therapy should autonomously commence as soon as possible after the onset of inflammation, should be controllable by a physician, and should not systemically block beneficial immune response in the long term. We describe a genetically encoded anti-inflammatory mammalian cell device based on a modular engineered genetic circuit comprising a sensor, an amplifier, a hresholder to restrict activation of a positive-feedback loop, a combination of advanced clinically used biopharmaceutical proteins, and orthogonal regulatory elements that linked modules into the functional device. This genetic circuit was autonomously activated by inflammatory signals, including endogenous cecal ligation and puncture (CLP)-induced inflammation in mice and serum from a systemic juvenile idiopathic arthritis (sIJA) patient, and could be reset externally by a chemical signal. The microencapsulated anti-inflammatory device significantly reduced the pathology in dextran sodium sulfate (DSS)-induced acute murine colitis, demonstrating a synthetic immunological approach for autonomous anti-inflammatory therapy.
Increasing the dynamic control space of mammalian transcription devices by combinatorial assembly of homologous regulatory elements from different bacterial species
78 Construction of mammalian gene regulation systems responsive to l-tryptophan. 78 Genetic elements originate from both Escherichia coli and Chlamydia trachomatis. 78 Novel combinatorial approach allow for new system characteristics to be obtained. 78 Various l-tryptophan analogues enable distinct gene response profiles. 78 Promoter, transactivator and inducer combinations enable system diversity.
The cumate gene-switch: a system for regulated expression in mammalian cells
pAbstract/p pBackground/p pA number of expression systems have been developed where transgene expression can be regulated. They all have specific characteristics making them more suitable for certain applications than for others. Since some applications require the regulation of several genes, there is a need for a variety of independent yet compatible systems./p pResults/p pWe have used the regulatory mechanisms of bacterial operons (itcmt /itand itcym/it) to regulate gene expression in mammalian cells using three different strategies. In the repressor configuration, regulation is mediated by the binding of the repressor (CymR) to the operator site (CuO), placed downstream of a strong constitutive promoter. Addition of cumate, a small molecule, relieves the repression. In the transactivator configuration, a chimaeric transactivator (cTA) protein, formed by the fusion of CymR with the activation domain of VP16, is able to activate transcription when bound to multiple copies of CuO, placed upstream of the CMV minimal promoter. Cumate addition abrogates DNA binding and therefore transactivation by cTA. Finally, an adenoviral library of cTA mutants was screened to identify a reverse cumate activator (rcTA), which activates transcription in the presence rather than the absence of cumate./p pConclusion/p pWe report the generation of a new versatile inducible expression system./p
Gas-inducible transgene expression in mammalian cells and mice
We describe the design and detailed characterization of a gas-inducible transgene control system functional in different mammalian cells, and prototype biopharmaceutical manufacturing. The -inducible -P() transactivator-promoter interaction of the -catabolizing regulon was engineered for gas-adjustable transgene expression in mammalian cells. Fungal retained its transactivation characteristics in a variety of mammalian cell lines and reversibly adjusted transgene from chimeric mammalian promoters (P(AIR)) containing P()-derived operators in a gaseous -dependent manner. implanted with microencapsulated cells engineered for -inducible regulation (AIR) of the secreted placental showed adjustable serum levels after exposure to different gaseous concentrations. AIR-controlled interferon-beta production in transgenic -K1-derived serum-free suspension cultures could be modulated by fine-tuning inflow and outflow of -containing gas during standard bioreactor operation. AIR technology could serve as a tool for therapeutic transgene dosing as well as biopharmaceutical manufacturing.
A novel mammalian expression system derived from components coordinating nicotine degradation in arthrobacter nicotinovorans pAO1
We describe the design and detailed characterization of 6-hydroxy-nicotine (6HNic)-adjustable transgene expression (NICE) systems engineered for lentiviral transduction and in vivo modulation of angiogenic responses. Arthrobacter nicotinovorans pAO1 encodes a unique catabolic machinery on its plasmid pAO1, which enables this Gram-positive soil bacterium to use the tobacco alkaloid nicotine as the exclusive carbon source. The 6HNic-responsive repressor-operator (HdnoR-ONIC) interaction, controlling 6HNic oxidase production in A.nicotinovorans pAO1, was engineered for generic 6HNic-adjustable transgene expression in mammalian cells. HdnoR fused to different transactivation domains retained its ONIC-binding capacity in mammalian cells and reversibly adjusted transgene transcription from chimeric ONIC-containing promoters (PNIC; ONIC fused to a minimal eukaryotic promoter [Pmin]) in a 6HNic-responsive manner. The combination of transactivators containing various transactivation domains with promoters differing in the number of operator modules as well as in their relative inter-ONIC and/or ONIC-Pmin spacing revealed steric constraints influencing overall NICE regulation performance in mammalian cells. Mice implanted with microencapsulated cells engineered for NICE-controlled expression of the human glycoprotein secreted placental alkaline phosphatase (SEAP) showed high SEAP serum levels in the absence of regulating 6HNic. 6HNic was unable to modulate SEAP expression, suggesting that this nicotine derivative exhibits control-incompatible pharmacokinetics in mice. However, chicken embryos transduced with HIV-1-derived self-inactivating lentiviral particles transgenic for NICE-adjustable expression of the human vascular endothelial growth factor 121 (VEGF121) showed graded 6HNic response following administration of different 6HNic concentrations. Owing to the clinically inert and highly water-soluble compound 6HNic, NICE-adjustable transgene control systems may become a welcome alternative to available drug-responsive homologs in basic research, therapeutic cell engineering and biopharmaceutical manufacturing.
Charpin-El Hamri G, et al. A synthetic multifunctional mammalian pH sensor and CO2 transgene-control device
Ausl盲nder et al. use a synthetic biology approach to engineer pH (CO2/H+)-sensitive transgene expression in mammalian cells. The pH-Sensor enabled CO2-programmable product-gene expression inside microfluidic devices and bioreactors and CO2-mediated control of logic gates. Designer cell implants providing blood pH-sensitive insulin expression corrected diabetic ketoacidosis in mice.
A synthe-tic mammalian gene circuit reveals antituberculosis compounds
Charpin-El-Hamri G, Fussenegger M. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice
Diet-induced is a lifestyle-associated medical condition that increases the risk of developing , and certain types of . Here we report the design of a closed-loop genetic circuit that constantly monitors blood fatty acid levels in the setting of diet-associated and coordinates reversible and adjustable expression of the clinically licensed appetite-suppressing pramlintide. Grafting of the proliferator-activated receptor- onto the -responsive repressor produces a synthetic -sensing receptor () that reversibly induces chimeric -specific promoters in a fatty acid-adjustable manner. with diet-induced in which microencapsulated cells engineered for -driven expression of pramlintide are implanted show significant reduction in food consumption, blood levels and body weight when put on a high-fat diet. Therapeutic designer circuits that monitor levels of pathologic metabolites and link these with the tailored expression of protein pharmaceuticals may provide new opportunities for the treatment of .
A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells
Abstract Synthetic biology has advanced the design of standardized transcription control devices that programme cellular behaviour. By coupling synthetic signalling cascade- and transcription factor-based gene switches with reverse and differential sensitivity to the licensed food additive vanillic acid, we designed a synthetic lineage-control network combining vanillic acid-triggered mutually exclusive expression switches for the transcription factors Ngn3 (neurogenin 3; OFF-ON-OFF) and Pdx1 (pancreatic and duodenal homeobox 1; ON-OFF-ON) with the concomitant induction of MafA (V-maf musculoaponeurotic fibrosarcoma oncogene homologue A; OFF-ON). This designer network consisting of different network topologies orchestrating the timely control of transgenic and genomic Ngn3, Pdx1 and MafA variants is able to programme human induced pluripotent stem cells (hIPSCs)-derived pancreatic progenitor cells into glucose-sensitive insulin-secreting beta-like cells, whose glucose-stimulated insulin-release dynamics are comparable to human pancreatic islets. Synthetic lineage-control networks may provide the missing link to genetically programme somatic cells into autologous cell phenotypes for regenerative medicine.
The food additive vanillic acid controls transgene expression in mammalian cells and mice
Trigger-inducible transcription-control devices that reversibly fine-tune transgene expression in response to molecular cues have significantly advanced the rational reprogramming of mammalian cells. When designed for use in future gene- and cell-based therapies the trigger molecules have to be carefully chosen in order to provide maximum specificity, minimal side-effects and optimal pharmacokinetics in a mammalian organism. Capitalizing on control components that enable Caulobacter crescentus to metabolize vanillic acid originating from lignin degradation that occurs in its oligotrophic freshwater habitat, we have designed synthetic devices that specifically adjust transgene expression in mammalian cells when exposed to vanillic acid. Even in mice transgene expression was robust, precise and tunable in response to vanillic acid. As a licensed food additive that is regularly consumed by humans via flavoured convenience food and specific fresh vegetable and fruits, vanillic acid can be considered as a safe trigger molecule that could be used for diet-controlled transgene expression in future gene- and cell-based therapies.
Synthetic biosensors for precise gene control and real-time monitoring of metabolites
http://nar.oxfordjournals.org/content/early/2015/07/07/nar.gkv616.short
Daoud-El Baba M, et al. Vitamin H-regulated transgene expression in mammalian cells
Although adjustable transgene expression systems are considered essential for future therapeutic and biopharmaceutical manufacturing applications, the currently available transcription control modalities all require side-effect-prone inducers such as immunosupressants, hormones and antibiotics for fine-tuning. We have designed a novel mammalian transcription-control system, which is reversibly fine-tuned by non-toxic vitamin H (also referred to as biotin). Ligation of vitamin H, by engineeredEscherichia colibiotin ligase (BirA), to a synthetic biotinylation signal fused to the tetracycline-dependent transactivator (tTA), enables heterodimerization of tTA to a streptavidin-linked transrepressor domain (KRAB), thereby abolishing tTA-mediated transactivation of specific target promoters. As heterodimerization of tTA to KRAB is ultimately conditional upon the presence of vitamin H, the system is vitamin H responsive. Transgenic Chinese hamster ovary cells, engineered for vitamin H-responsive gene expression, showed high-level, adjustable and reversible production of a human model glycoprotein in bench-scale culture systems, bioreactor-based biopharmaceutical manufacturing scenarios, and after implantation into mice. The vitamin H-responsive expression systems showed unique band pass filter-like regulation features characterized by high-level expression at low (0~2 nM biotin), maximum repression at intermediate (100~1000 nM biotin), and high-level expression at increased (>100 000 nM biotin) biotin concentrations. Sequential ON-to-OFF-to-ON, ON-to-OFF and OFF-to-ON expression profiles with graded expression transitions can all be achieved by simply increasing the level of a single inducer molecule without exchanging the culture medium. These novel expression characteristics mediated by an FDA-licensed inducer may foster advances in therapeutic cell engineering and manufacturing of difficult-to-produce protein therapeutics.
A biotin-triggered genetic switch in mammalian cells and mice
Adjustable and reversible transgene expression systems enabling precise control of metabolic pathways and tunable production of specific target proteins have been essential for conditional reprogramming of mammalian cells to achieve progress in basic and applied bioengineering disciplines. Most of the currently available transgene control modalities have been designed to be responsive to clinically licensed pharmacologically active drugs which were expected to prevail in future clinical trials yet raised concerns about side effects when administered long term at subclinical doses. We have chosen vitamin H, also known as biotin, to control target gene transcription in mammalian cells in a potentially side effect-free manner. BirA, the Escherichia coli repressor of the biotin biosynthesis operon, was fused to the Herpes simplex transactivation domain to generate a biotin-dependent transactivator (BIT), which, in the presence of biotin, binds and activates chimeric target promoters (P BIT) harboring BirA-specific operator sites 5 of a minimal promoter. Biotin-inducible transgene expression was functional in a variety of rodent, monkey and human cell lines, showed excellent adjustability and reversibility in transgenic Chinese hamster ovary cell lines, provided precise product gene control in standard bioreactor cultures and enabled dose-dependent vitamin H control of a human glycoprotein in mice. The combination of a side effect-free inducer, precise and reversible transcription tunability and broad functionality in different cell types as well as in entire animals represents a unique asset for the use of biotin-inducible transgene control in future gene therapy, tissue engineering and biopharmaceutical manufacturing scenarios.