Chinese Medical Sciences Journal ›› 2024, Vol. 39 ›› Issue (1): 19-28.doi: 10.24920/004338
Previous Articles Next Articles
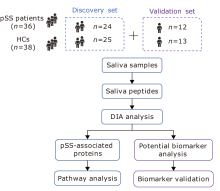
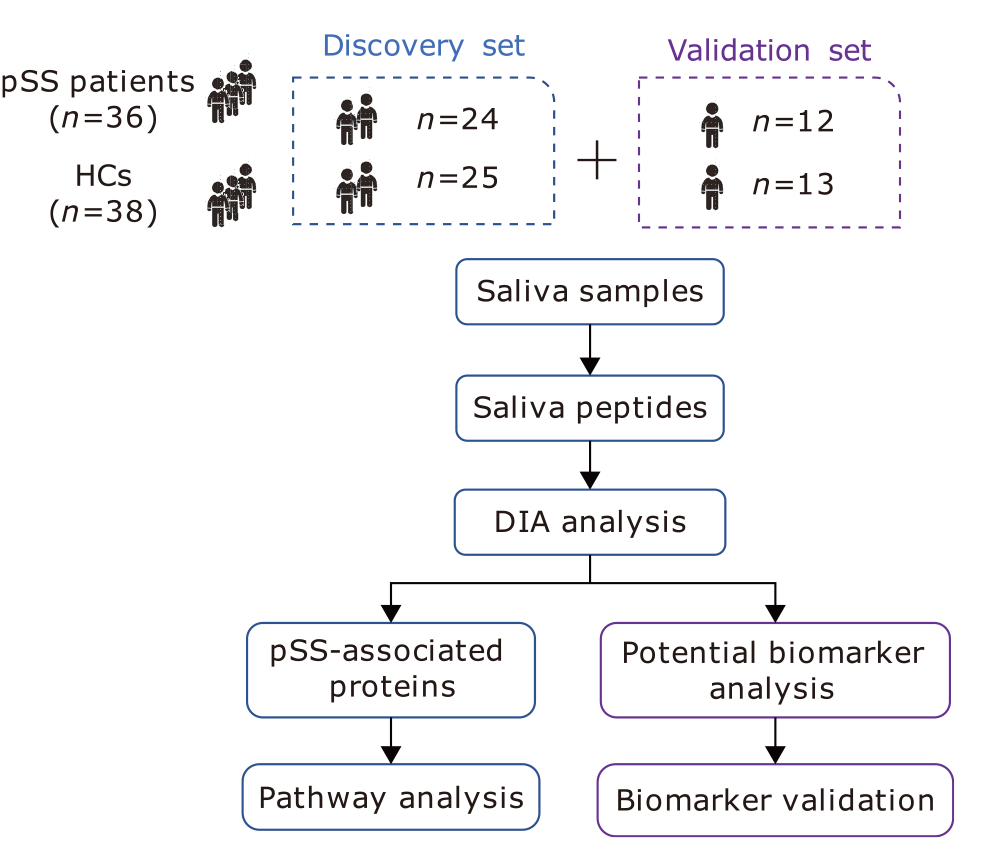
Data-Independent Acquisition-Based Quantitative Proteomic Analysis Reveals Potential Salivary Biomarkers of Primary Sjögren’s Syndrome
Yi-Chao Tian1, Chun-Lan Guo1, Zhen Li1, Xin You2, Xiao-Yan Liu3, Jin-Mei Su2, Si-Jia Zhao1, Yue Mu1, Wei Sun3, *( ), Qian Li1, *(
), Qian Li1, *( )
)
- 1Department of Stomatology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, China
2Department of Rheumatology and Clinical Immunology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, China
3Proteomics Center, Core Facility of Instrument, Institute of Basic Medical Sciences of Chinese Academy of Medical Sciences & School of Basic Medicine Peking Union Medical College, Beijing 100730, China
-
Received:2024-01-15Accepted:2024-03-08Published:2024-03-31 -
Contact:* Qian Li E-mail:liqian4230@pumch.cn ;Wei Sun, E-mail:sunwei@ibms.pumc.edu.cn .
Cite this article
Yi-Chao Tian, Chun-Lan Guo, Zhen Li, Xin You, Xiao-Yan Liu, Jin-Mei Su, Si-Jia Zhao, Yue Mu, Wei Sun, Qian Li. Data-Independent Acquisition-Based Quantitative Proteomic Analysis Reveals Potential Salivary Biomarkers of Primary Sjögren’s Syndrome[J].Chinese Medical Sciences Journal, 2024, 39(1): 19-28.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1.
Baseline characteristics of pSS patients and healthy controls"
| Characteristics | pSS group | HCs group | t/X2 | P value | |||
|---|---|---|---|---|---|---|---|
| Discovery (n=24) | Validation (n=12) | Discovery (n=25) | Validation (n=13) | ||||
| Age, yrs, mean±SD | 41.1±10.9 | 40.6±12.8 | 41.4±12.2 | 44.1±12.7 | 0.049 | 0.88 | |
| Sex, females [n(%)] | 21(87.5) | 10(83.3) | 23 (92) | 12 (92.3) | 0.075 | 0.60 | |
| Anti-SSA/Ro antibody + [n(%)] | 16(66.7) | 9(75.0) | / | / | 0.574 | 0.682 | |
| Anti-SSB/La antibody + [n(%)] | 3(12.5) | 1(8.3) | / | / | 0.112 | 1 | |
| Immunoglobulin G + [n(%)] | 12(50.0) | 6(50.0) | / | / | 0 | 1 | |
| Labial gland biopsy + [n(%)] | 13(54.2) | 9(75.0) | / | / | 2.595 | 0.205 | |
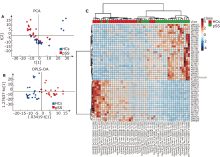
Figure 3.
Proteomic profiling of pSS and HCs. (A) PCA score plot of proteomics profiling btween pSS and HCs. (B) OPLS-DA score plot of proteomics profiling between pSS and HCs. (C) Heatmap of the relative intensity of the top 50 (P value) differentially expressed proteins between pSS and HCs."
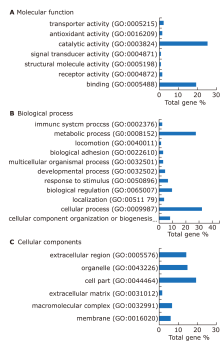
Figure 4.
Expression profile and functional annotation of the significantly differential proteins. (A) GO analysis performed for the differential proteins of pSS in terms of molecular function. (B) GO analysis performed for the differential proteins of pSS in terms of biological process. (C) GO analysis performed for the differential proteins of pSS in terms of cellular component. Categories with a constitution of at least 2% are displayed in the bar charts."
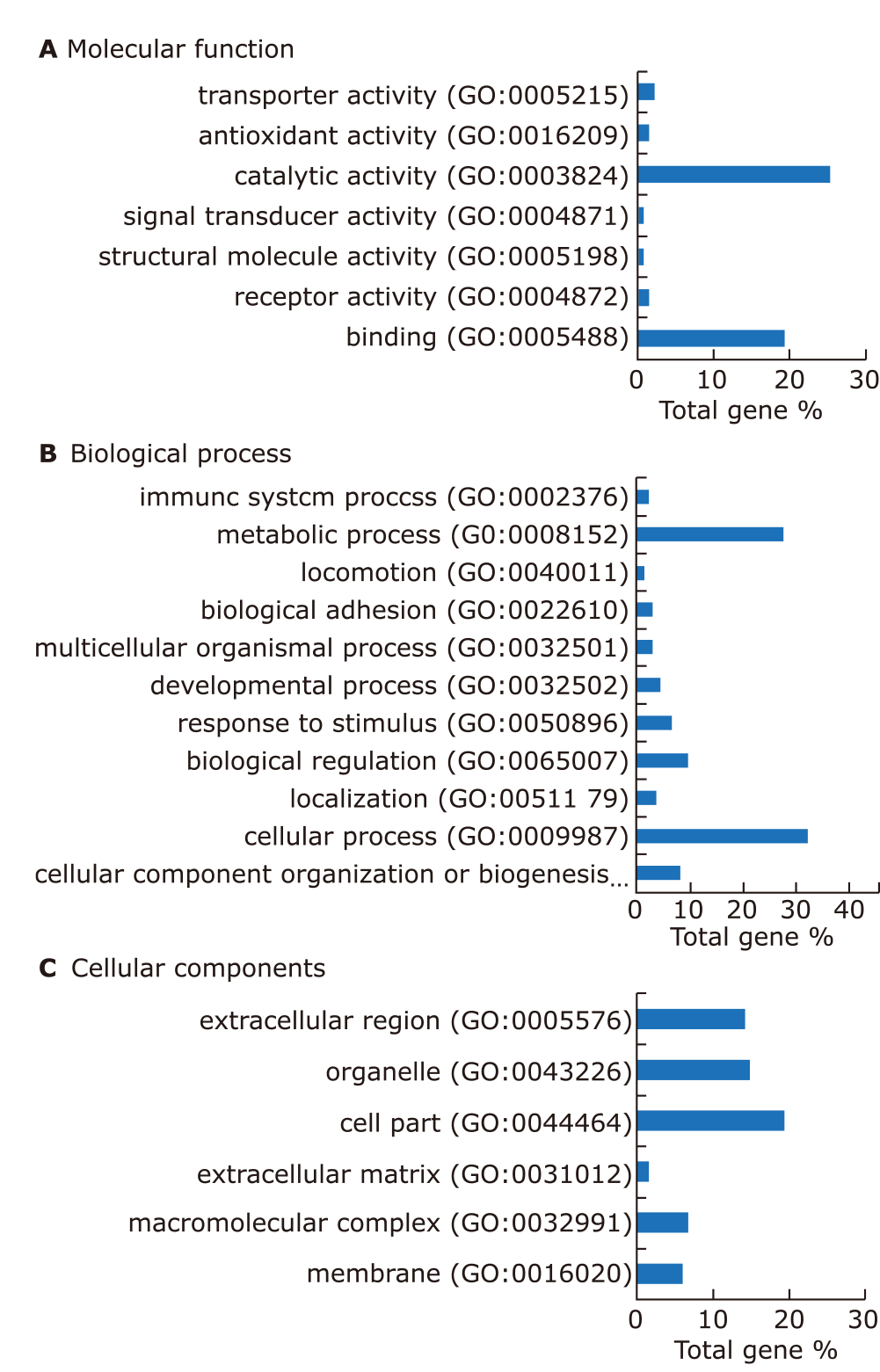
Figure 5.
Pathway and function analysis of differential proteins by IPA. (A) Top 10 pathways enriched by the 136 differential proteins. (B) Top 10 functions enriched by the 136 differential proteins. (C) Potential protein-protein interaction networks involved in regulate and control inflammation, immune response, cellular death and survival. Lines between the proteins indicates direct (—) or indirect (- - -) interactions; pink nodes: proteins that up regulated in SS significantly; green nodes: proteins that down regulated in SS significantly. Numbers below the nodes indicates the fold changes of proteins between SS and control. node shapes represent protein classes."
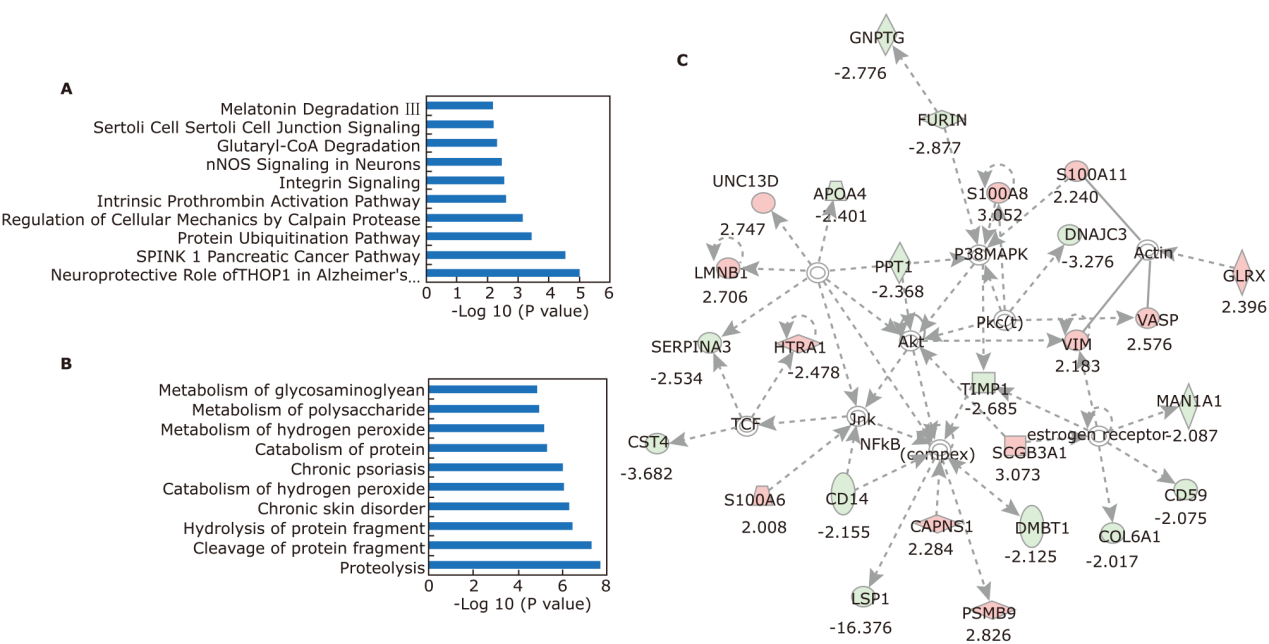
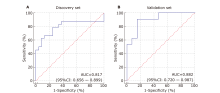
Table 2.
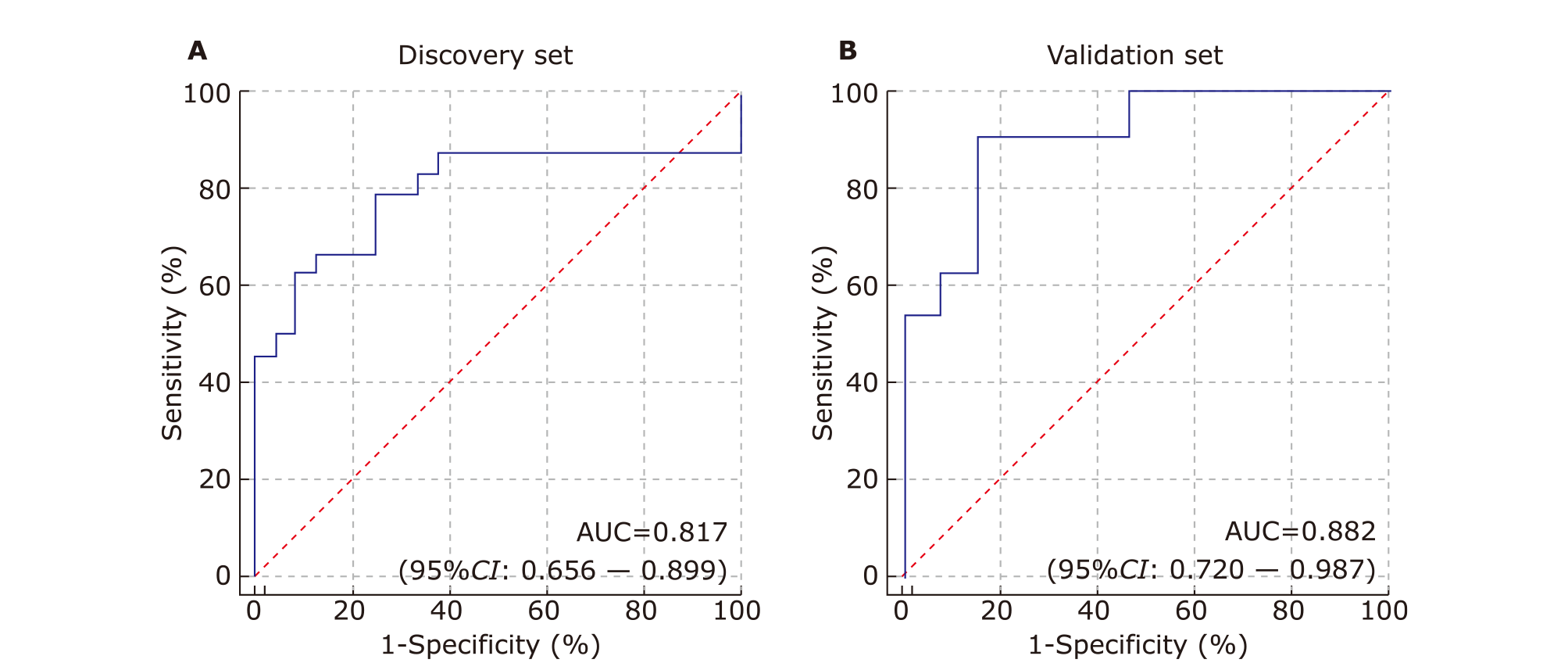
Diagnostic performance of differentially expressed proteins in distinction of pSS patients from healthy controls using saliva proteomics"
| No. | Protein name | AUC | P value | Fold change |
|---|---|---|---|---|
| 1 | Q13217 | 0.86285 | 0.002438 | 2.0129 |
| 2 | P01034 | 0.85764 | 0.006314 | 2.6949 |
| 3 | P07711 | 0.85243 | 0.001923 | 1.831 |
| 4 | P09958 | 0.85069 | 0.001031 | 1.8654 |
| 5 | P28065 | 0.85069 | 7.72E-06 | -1.2467 |
| 6 | O43852 | 0.84201 | 6.80E-04 | 1.4389 |
| 7 | O43505 | 0.83854 | 2.39E-04 | 1.4676 |
| 8 | P25789 | 0.82812 | 9.41E-05 | -1.276 |
| 9 | Q9UBX7 | 0.81771 | 0.00464 | 1.3578 |
| 10 | Q9NRJ3 | 0.81597 | 0.024021 | 1.4358 |
| 11 | P35030 | 0.81424 | 0.001265 | 1.3919 |
| 12 | P12109 | 0.8125 | 0.008958 | 1.3342 |
| 13 | O75976 | 0.80903 | 6.50E-04 | 1.3292 |
| 14 | Q92743 | 0.80035 | 0.46752 | 1.0645 |
| 15 | P16870 | 0.7934 | 0.001147 | 1.3544 |
| 16 | P13798 | 0.78646 | 5.01E-04 | -1.0262 |
| 17 | P01011 | 0.75347 | 0.018898 | 2.5295 |
| 18 | P32320 | 0.74306 | 0.001919 | -0.88055 |
| 19 | P08670 | 0.73785 | 0.006534 | -1.581 |
| 1. | González S, Sung H, Sepúlveda D, et al. Oral manifestations and their treatment in Sjögren’s syndrome. Oral Dis 2014; 20: 153-61. doi: 10.1111/odi.12105. |
| 2. | Brito-Zerón P, Baldini C, Bootsma H, et al. Sjögren syndrome. Nat Rev Dis Primers 2016; 2: 16047. doi: 10.1016/j.medcli.2022.10.007. |
| 3. | Soret P le Dantec C, Desvaux E, et al. A new molecular classification to drive precision treatment strategies in primary Sjögren’s syndrome. Nat Commun 2021; 12: 3523. doi: 10.1038/s41467-021-23472-7. |
| 4. |
Vitorino R, Lobo MJC, Ferrer-Correira AJ, et al. Identification of human whole saliva protein components using proteomics. Proteomics 2004; 4: 1109-15. doi:10.1002/pmic.200300638.
pmid: 15048992 |
| 5. |
Malamud D. Saliva as a diagnostic fluid. BMJ 1992; 305: 207-8. doi:10.1016/j.cden.2010.08.004.
pmid: 1290500 |
| 6. |
Loo J A, Yan W, Ramachandran P, et al. Comparative human salivary and plasma proteomes. J Dent Res 2010; 89: 1016-23. doi:10.1177/0022034510380414.
pmid: 20739693 |
| 7. | Hu S, Vissink A, Arellano M, et al. Identification of autoantibody biomarkers for primary Sjögren’s syndrome using protein microarrays. Proteomics 2011; 11: 1499-507. doi:10.1002/pmic.201000206. |
| 8. |
Deutsch O, Krief G, Konttinen YT, et al. Identification of Sjögren’s syndrome oral fluid biomarker candidates following high-abundance protein depletion. Rheumatology (Oxford) 2015; 54: 884-90. doi: 10.1093/rheumatology/keu405.
pmid: 25339641 |
| 9. | Katsiougiannis S, Wong DTW. The proteomics of saliva in Sjögren’s syndrome. Rheum Dis Clin North Am 2016; 42: 449-56. doi:10.1016/j.rdc.2016.03.004. |
| 10. | Jonsson R, Brokstad KA, Jonsson MV, et al. Current concepts on Sjögren’s syndrome - classification criteria and biomarkers. Eur J Oral Sci 2018; 126 Suppl 1: 37-48. doi: 10.1111/eos.12536. |
| 11. |
Ferraccioli G, de Santis M, Peluso G, et al. Proteomic approaches to Sjögren’s syndrome: a clue to interpret the pathophysiology and organ involvement of the disease. Autoimmun Rev 2010; 9: 622-6. doi:10.1016/j.autrev.2010.05.010.
pmid: 20462525 |
| 12. |
Baldini C, Giusti L, Bazzichi L, et al. Proteomic analysis of the saliva: a clue for understanding primary from secondary Sjögren’s syndrome? Autoimmun Rev 2008; 7: 185-91. doi: 10.1016/j.autrev.2007.11.002.
pmid: 18190876 |
| 13. | Chen WQn, Cao H, Lin J, et al. Biomarkers for primary Sjögren’s syndrome. Genomics Proteomics Bioinformatics 2015; 13: 219-23. doi: 10.2217/bmm-2017-0297. |
| 14. |
Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13: 227-32. doi: 10.1038/nrg3185.
pmid: 22411467 |
| 15. |
Callister SJ, Barry RC, Adkins JN, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res 2006; 5: 277-86. doi: 10.1021/pr050300l.
pmid: 16457593 |
| 16. | Maddali BS, Campana G, D’Agata A, et al. The diagnosis value of beta 2-microglobulin and immunoglobulins in primary Sjögren’s syndrome. Clin Rheumatol 1995; 14: 151-6. doi: 10.1007/BF02214934. |
| 17. |
Baldini C, Gallo A, Perez P, et al. Saliva as an ideal milieu for emerging diagnostic approaches in primary Sjögren’s syndrome. Clin Exp Rheumatol 2012; 30: 785-90.
pmid: 23009763 |
| 18. | Jin YB, Dai YJ, Chen JL, et al. Anti-carbonic anhydrase II antibody reflects urinary acidification defect especially in proximal renal tubules in patients with primary Sjögren syndrome. Medicine (Baltimore) 2023; 102: e32673. doi: 10.1097/MD.0000000000032673. |
| 19. | Qin BD, Wang JQ, Yang ZX, et al. Epidemiology of primary Sjögren’s syndrome: a systematic review and meta-analysis. Ann Rheum Dis 2015; 74: 1983-9. doi: 10.1136/annrheumdis-2014-205375. |
| 20. | Goules AV, Tzioufas AG. Primary Sjögren’s syndrome: clinical phenotypes, outcome and the development of biomarkers. Immunol Res 2017; 65: 331-344. doi: 10.1007/s12026-016-8844-4. |
| 21. | Chaudhury NMA, Proctor GB, Karlsson NG, et al. Reduced mucin-7 (Muc7) sialylation and altered saliva rheology in sjögren’s syndrome associated oral dryness. Mol Cell Proteomics 2016; 15: 1048-59. doi:10.1074/mcp.M115.052993. |
| 22. | Horvatovich P, Végvári Á, Saul J, et al. In vitro transcription/translation system: a versatile tool in the search for missing proteins. J Proteome Res 2015; 14: 3441-51. doi:10.1021/acs.jproteome.5b00486. |
| 23. | Meyer JG, D’Souza AK, Sorensen DJ, et al. Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J Am Soc Mass Spectrom 2016; 27: 1758-1771. doi:10.1007/s13361-016-1476-z. |
| 24. | Gao TG, Lou CX, Wang Y, et al. Anti-carbonic anhydrase goldbodies by conformational reconstruction of the complementary-determining regions of phage-displayed antibodies. ChemMedChem 2023; 18: e202300185. doi:10.1002/cmdc.202300185. |
| 25. | Guo ZG, Liu XJ, Li ML, et al. Differential urinary glycoproteome analysis of type 2 diabetic nephropathy using 2D-LC-MS/MS and iTRAQ quantification. J Transl Med 2015; 13: 371. doi: 10.1186/s12967-015-0712-9. |
| 26. | Xiao XP, Liu YR, Guo ZG, et al. Comparative proteomic analysis of the influence of gender and acid stimulation on normal human saliva using LC/MS/MS. Proteomics Clin Appl 2017; 11: undefined. doi: 10.1002/prca.201600142. |
| Garza-García F, Delgado-García G, Garza-Elizondo M, et al. Salivary β2-microglobulin positively correlates with ESSPRI in patients with primary Sjögren’s syndrome. Rev Bras Reumatol Engl Ed 2017; 57: 182-4. doi: 10.1016/j.rbre.2016.11.001. | |
| 27. | Thatayatikom A, Jun I, Bhattacharyya I, et al. The diagnostic performance of early Sjögren’s syndrome autoantibodies in juvenile Sjögren’s syndrome: The University of Florida Pediatric Cohort study. Front Immunol 2021; 12: 704193. doi:10.3389/fimmu.2021.704193. |
| 28. | Margaretten M. Neurologic manifestations of primary Sjögren syndrome. Rheum Dis Clin North Am 2017; 43: 519-29. doi: 10.1016/j.rdc.2017.06.002. |
| 29. | Silva LM, Clements JA. Mass spectrometry based proteomics analyses in kallikrein-related peptidase research: implications for cancer research and therapy. Expert Rev Proteomics 2017; 14: 1119-30. doi:10.1080/14789450.2017.1389637. |
| 30. | Botrè F, Botrè C, Podestà E, et al. Effect of anti-carbonic anhydrase antibodies on carbonic anhydrases I and II. Clin Chem 2003; 49: 1221-3. doi: 10.1373/49.7.1221. |
| 31. |
Makino K, Jinnin M, Makino T, et al. Serum levels of soluble carbonic anhydrase IX are decreased in patients with diffuse cutaneous systemic sclerosis compared to those with limited cutaneous systemic sclerosis. Biosci Trends 2014; 8: 144-8. doi: 10.5582/bst.2014.01020.
pmid: 25030848 |
| 32. |
Aparisi L, Farre A, Gomez-Cambronero L, et al. Antibodies to carbonic anhydrase and IgG4 levels in idiopathic chronic pancreatitis: relevance for diagnosis of autoimmune pancreatitis. Gut 2005; 54: 703-9. doi: 10.1136/gut.2004.047142.
pmid: 15831920 |
| 33. |
Invernizzi P, Selmi C, Zuin M, et al. Lack of serum antibodies to membrane bound carbonic anhydrase IV in patients with primary biliary cirrhosis. Gut 2005; 54: 1665. doi: 10.1136/gut.2005.072389.
pmid: 16227372 |
| 34. | Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012; 11: O111.016717. doi: 10.1074/mcp.O111.016717. |
| [1] | Changyi Liu, Xiaoqing Liu, Xiaochun Shi. Clinical Features of Spontaneous Remission in the Classic Fever of Unknown Origin: A Retrospective Study [J]. Chinese Medical Sciences Journal, 2022, 37(2): 134-141. |
| [2] | Huizi Gong, Mengyin Wu, Jun Li, Heyi Zheng. The Great Imitator: Atypical Cutaneous Manifestations of Primary Syphilitic Chancre [J]. Chinese Medical Sciences Journal, 2021, 36(4): 279-283. |
| [3] | Pengfei Qu, Baoliang Bai, Ting Duan, Kai Liu, Jinliang Du, Xin Xiong, Penglin Jia, Zhongchun Sun, Puping Lei. Pneumonia, Multiple Pulmonary Infarction and Abscess Caused by a Bamboo Stick Accidentally Piercing into Chest: a Case Misdiagnosed as Pulmonary Tuberculosis [J]. Chinese Medical Sciences Journal, 2021, 36(3): 252-256. |
| [4] | Lianyan Xu, Ke Yan, Le Lu, Weihong Zhang, Xu Chen, Xiaofei Huo, Jingjing Lu. External and Internal Validation of a Computer Assisted Diagnostic Model for Detecting Multi-Organ Mass Lesions in CT images [J]. Chinese Medical Sciences Journal, 2021, 36(3): 210-217. |
| [5] | Dasheng Li,Dawei Wang,Nana Wang,Haiwang Xu,He Huang,Jianping Dong,Chen Xia. An Insight of the First Community Infected COVID-19 Patient in Beijing by Imported Case: Role of Deep Learning-Assisted CT Diagnosis [J]. Chinese Medical Sciences Journal, 2021, 36(1): 66-71. |
| [6] | Wang Xiaolei, Meng Shanshan, Duan Kehang, Hu Yaowei, Wei Feng. Treatment of Retroperitoneal Cavernous Lymphangioma: A Case Report [J]. Chinese Medical Sciences Journal, 2020, 35(3): 283-285. |
| [7] | Wu Ziquan, Zeng Delu, Yao Jiangling, Bian Yangyang, Gu Yuntao, Meng Zhulong, Fu Jian, Peng Lei. Research Progress on Diagnosis and Treatment of Chronic Osteomyelitis [J]. Chinese Medical Sciences Journal, 2019, 34(3): 211-220. |
| [8] | Wang Yingwei, Zhang Xinghua, Wang Botao, Wang Ye, Liu Mengqi, Wang Haiyi, Ye Huiyi, Chen Zhiye. Value of Texture Analysis of Intravoxel Incoherent Motion Parameters in Differential Diagnosis of Pancreatic Neuroendocrine Tumor and Pancreatic Adenocarcinoma [J]. Chinese Medical Sciences Journal, 2019, 34(1): 1-9. |
| [9] | Wang Botao, Liu Mingxia, Chen Zhiye. Differential Diagnostic Value of Texture Feature Analysis of Magnetic Resonance T2 Weighted Imaging between Glioblastoma and Primary Central Neural System Lymphoma [J]. Chinese Medical Sciences Journal, 2019, 34(1): 10-17. |
| [10] | Wang Botao, Fan Wenping, Xu Huan, Li Lihui, Zhang Xiaohuan, Wang Kun, Liu Mengqi, You Junhao, Chen Zhiye. Value of Magnetic Resonance Imaging Texture Analysis in the Differential Diagnosis of Benign and Malignant Breast Tumors [J]. Chinese Medical Sciences Journal, 2019, 34(1): 33-37. |
| [11] | Bai Mingjian, Feng Jing, Liang Guowei. Urinary Myeloperoxidase to Creatinine Ratio as a New Marker for Diagnosis of Urinary Tract Infection [J]. Chinese Medical Sciences Journal, 2018, 33(3): 152-159. |
| [12] | Chen Zhiye, Liu Mengqi, Yu Shengyuan, Ma Lin. Multi-parametric MRI Diagnoses Cerebellar Hemangioblastoma: A Case Report [J]. Chinese Medical Sciences Journal, 2018, 33(3): 188-193. |
| [13] | Dong Dexin, Ji Zhigang, Li Hanzhong, Yan Weigang, Zhang Yushi. Preliminary Application of WCX Magnetic Bead-Based Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Analyzing the Urine of Renal Clear Cell Carcinoma [J]. Chinese Medical Sciences Journal, 2017, 32(4): 248-252. |
| [14] | Li Tao, Zhao Shaohong, Li Jinfeng, Huang Zili, Luo Chuncai, Yang Li. Value of Multi-detector CT in Detection of Isolated Spontaneous Superior Mesenteric Artery Dissection [J]. Chinese Medical Sciences Journal, 2017, 32(1): 28-33. |
| [15] | Ai-chun Liu, Yan-ying Liu, Yan Li, Li Zhang, Zhan-guo Li. Multiple Myeloma Mimicking Spondyloarthritis: a Case Report [J]. Chinese Medical Sciences Journal, 2014, 29(4): 245-247. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
|