Chinese Medical Sciences Journal ›› 2022, Vol. 37 ›› Issue (1): 31-43.doi: 10.24920/003966
• Original Article • Previous Articles Next Articles
Identifying and Validating a Novel miRNA-mRNA Regulatory Network Associated with Prognosis in Lung Adenocarcinoma
Wenqin Xu1, 2, Jingjing Ye1, 2, Tianbing Chen1, 2, *( )
)
- 1Central Laboratory, Yijishan Hospital of Wannan Medical College, Wuhu, Anhui 241002, China
2Key Laboratory of Non-coding RNA Transformation Research of Anhui Higher Education Institution, Wannan Medical College, Wuhu, Anhui 241002, China
-
Received:2021-07-08Accepted:2021-11-30Published:2022-03-31Online:2022-03-08 -
Contact:Tianbing Chen E-mail:tianbingchen1987@wnmc.edu.cn
Cite this article
Wenqin Xu, Jingjing Ye, Tianbing Chen. Identifying and Validating a Novel miRNA-mRNA Regulatory Network Associated with Prognosis in Lung Adenocarcinoma[J].Chinese Medical Sciences Journal, 2022, 37(1): 31-43.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
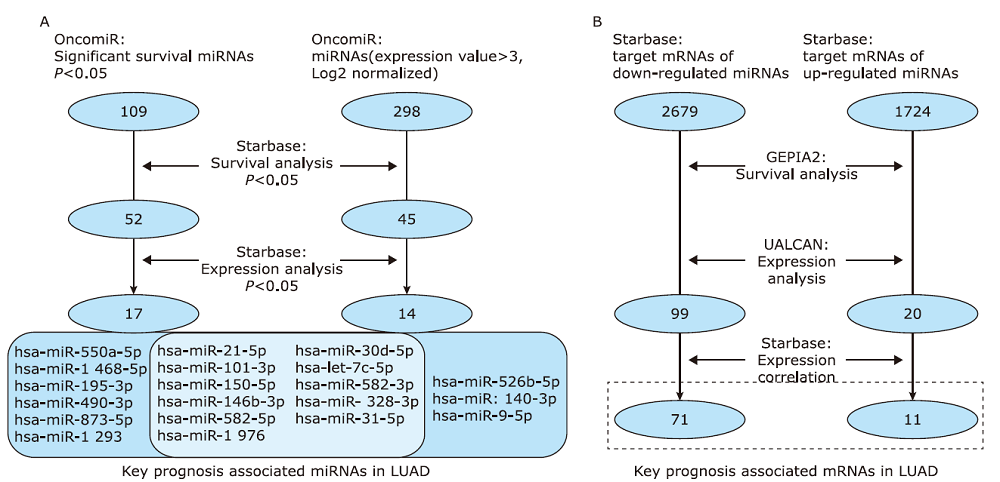
Table 1
Expression details and prognostic values of key miRNAs in LUAD"
| miRNA name | miRNA type | Expression details | Prognostic value | |||||
|---|---|---|---|---|---|---|---|---|
| Cancer Exp | Normal Exp | Fold Change | P Value | Hazard Ration | log-Rank P | |||
| hsa-miR-328-3p | Down-regulated / good prognosis | 36.93 | 674.10 | 0.05 | 0.0002 | 0.69 | 0.014 | |
| hsa-miR-1976 | Down-regulated / good prognosis | 14.73 | 114.84 | 0.13 | 6E-11 | 0.66 | 0.0053 | |
| hsa-let-7c-5p | Down-regulated / good prognosis | 1258.23 | 5235.5 | 0.24 | 2E-18 | 0.74 | 0.043 | |
| hsa-miR-146b-3p | Down-regulated / good prognosis | 227.52 | 966.56 | 0.24 | 0.0002 | 0.74 | 0.045 | |
| hsa-miR-150-5p | Down-regulated / good prognosis | 1108.92 | 3574.06 | 0.31 | 0.0084 | 0.74 | 0.046 | |
| hsa-miR-140-3p | Down-regulated / good prognosis | 908.08 | 2111.63 | 0.43 | 6E-15 | 0.73 | 0.035 | |
| hsa-miR-30d-5p | Down-regulated / good prognosis | 13296.36 | 23797.41 | 0.56 | 2E-05 | 0.62 | 0.0017 | |
| hsa-miR-101-3p | Down-regulated / good prognosis | 11939.12 | 14362.07 | 0.83 | 1E-09 | 0.65 | 0.0038 | |
| hsa-miR-490-3p | Down-regulated / good prognosis | 0.57 | 2.69 | 0.21 | 3E-09 | 0.72 | 0.033 | |
| hsa-miR-195-3p | Down-regulated / good prognosis | 3.42 | 13.21 | 0.26 | 3E-06 | 0.59 | 0.00046 | |
| hsa-miR-1468-5p | Down-regulated / good prognosis | 4.66 | 9.25 | 0.5 | 0.0026 | 0.64 | 0.0034 | |
| hsa-miR-526b-5p | Down-regulated / good prognosis | 7.11 | 11.19 | 0.64 | 2E-07 | 1.50 | 0.0093 | |
| hsa-miR-582-3p | Up-regulated / poor prognosis | 440.1 | 126.36 | 3.48 | 5E-07 | 1.36 | 0.039 | |
| hsa-miR-21-5p | Up-regulated / poor prognosis | 358144.55 | 38949.90 | 9.20 | 6E-101 | 1.45 | 0.014 | |
| hsa-miR-9-5p | Up-regulated / poor prognosis | 3568.81 | 35.32 | 101.04 | 3E-19 | 1.49 | 0.0079 | |
| hsa-miR-550a-5p | Up-regulated / poor prognosis | 4.77 | 3.24 | 1.47 | 0.016 | 1.50 | 0.0067 | |
| hsa-miR-582-5p | Up-regulated / poor prognosis | 20.23 | 6.00 | 3.37 | 5E-05 | 1.52 | 0.0051 | |
| hsa-miR-873-5p | Up-regulated / poor prognosis | 0.47 | 0.06 | 8.40 | 0.037 | 1.57 | 0.0031 | |
| hsa-miR-1293 | Up-regulated / poor prognosis | 0.30 | 0.01 | 30.34 | 0.0065 | 1.88 | 0.000025 | |
| hsa-miR-31-5p | Up-regulated / poor prognosis | 17.01 | 0.42 | 40.94 | 1E-08 | 1.39 | 0.032 | |
Table 2
Expression details and prognostic values of key mRNAs in LUAD"
| No. | Gene type | Gene Name | Expression details (Starbase) | Prognostic value (GEPIA2) | No. | Gene type | Gene Name | Expression details (Starbase) | Prognostic value (GEPIA2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer Exp | Normal Exp | Fold Change | P Value | Log rank P | Cancer Exp | Normal Exp | Fold Change | P Value | Log rank P | |||||||||
| 1 | Onco-mRNA | ESPL1 | 1.89 | 0.2 | 9.36 | 1.50E-44 | 0.0023 | 22 | Onco-mRNA | DDIT4 | 67.27 | 28.83 | 2.33 | 3.80E-08 | 0.0017 | |||
| 2 | Onco-mRNA | CCNB2 | 7.76 | 0.86 | 9 | 3.70E-56 | 0.00079 | 23 | Onco-mRNA | AHCY | 46.38 | 22.08 | 2.1 | 1.70E-21 | 0.0049 | |||
| 3 | Onco-mRNA | RRM2 | 11.64 | 1.12 | 10.37 | 1.40E-53 | 1.5e-05 | 24 | Onco-mRNA | RACGAP1 | 8.61 | 2.77 | 3.11 | 1.10E-22 | 0.00062 | |||
| 4 | Onco-mRNA | FANCI | 4.32 | 0.99 | 4.37 | 2.30E-43 | 0.043 | 25 | Onco-mRNA | CDCA4 | 8.52 | 2.84 | 3 | 1.60E-29 | 0.00081 | |||
| 5 | Onco-mRNA | YWHAZ | 120.83 | 61.68 | 1.96 | 1.30E-26 | 0.00023 | 26 | Onco-mRNA | CDCA8 | 8.67 | 0.95 | 9.16 | 2.50E-59 | 0.0026 | |||
| 6 | Onco-mRNA | GARS | 34.42 | 15.05 | 2.29 | 1.90E-34 | 0.00018 | 27 | Onco-mRNA | DNAJC9 | 4.59 | 2.3 | 1.99 | 2.10E-28 | 0.00057 | |||
| 7 | Onco-mRNA | EIF4A3 | 19.96 | 11.41 | 1.75 | 1.20E-18 | 0.013 | 28 | Onco-mRNA | UCK2 | 4.75 | 1.76 | 2.7 | 5.50E-09 | 0.00012 | |||
| 8 | Onco-mRNA | RHOV | 19.46 | 1.56 | 12.48 | 1.20E-26 | 0.0023 | 29 | Onco-mRNA | SMUG1 | 7.13 | 3.22 | 2.21 | 7.20E-38 | 0.004 | |||
| 9 | Onco-mRNA | LDHA | 141.93 | 51.1 | 2.78 | 1.20E-37 | 3e-05 | 30 | Onco-mRNA | PDIA6 | 76.66 | 33.9 | 2.26 | 6.90E-26 | 0.005 | |||
| 10 | Onco-mRNA | DIABLO | 1.77 | 1.09 | 1.63 | 3.00E-19 | 0.00058 | 31 | Onco-mRNA | CACYBP | 17.91 | 9.02 | 1.98 | 2.70E-27 | 0.00041 | |||
| 11 | Onco-mRNA | MRPL12 | 24.41 | 9.6 | 2.54 | 1.20E-14 | 5e-04 | 32 | Onco-mRNA | INTS7 | 6.82 | 3.39 | 2.01 | 7.70E-31 | 1.3e-05 | |||
| 12 | Onco-mRNA | POLR2D | 9.33 | 5.24 | 1.78 | 7.70E-33 | 0.0074 | 33 | Onco-mRNA | SLC7A11 | 5.03 | 0.55 | 9.14 | 7.40E-18 | 0.036 | |||
| 13 | Onco-mRNA | CALU | 66.43 | 30.23 | 2.2 | 4.90E-21 | 0.016 | 34 | Onco-mRNA | MRPS10 | 26.05 | 14.3 | 1.82 | 1.70E-24 | 0.01 | |||
| 14 | Onco-mRNA | NME4 | 25.98 | 9.02 | 2.88 | 6.20E-30 | 0.014 | 35 | Onco-mRNA | SLC2A1 | 38.71 | 3.14 | 12.33 | 1.80E-45 | 2.4e-05 | |||
| 15 | Onco-mRNA | PMAIP1 | 10.42 | 3.01 | 3.46 | 7.50E-13 | 0.0015 | 36 | Onco-mRNA | BIRC5 | 11.5 | 0.82 | 14.08 | 3.90E-55 | 0.00022 | |||
| 16 | Onco-mRNA | RFC2 | 15.71 | 8.11 | 1.94 | 1.70E-19 | 0.031 | 37 | Onco-mRNA | TTLL12 | 13.47 | 6.48 | 2.08 | 1.50E-15 | 0.00099 | |||
| 17 | Onco-mRNA | IGF2BP3 | 1.84 | 0.11 | 16.91 | 1.60E-14 | 0.0017 | 38 | Onco-mRNA | TIMM50 | 9.31 | 5.16 | 1.81 | 1.50E-15 | 0.03 | |||
| 18 | Onco-mRNA | KPNA2 | 36.71 | 8.42 | 4.36 | 3.10E-38 | 0.00038 | 39 | Onco-mRNA | CKS1B | 13.34 | 3.69 | 3.62 | 1.90E-32 | 4.4e-05 | |||
| 19 | Onco-mRNA | KIF2C | 7.05 | 0.54 | 13.11 | 1.60E-58 | 0.016 | 40 | Onco-mRNA | GPN1 | 13.77 | 8.2 | 1.68 | 7.20E-19 | 0.005 | |||
| 20 | Onco-mRNA | BZW1 | 20.71 | 11.81 | 1.75 | 5.00E-17 | 0.012 | 41 | Onco-mRNA | RAD51 | 2.43 | 0.49 | 4.98 | 7.10E-40 | 0.0026 | |||
| 21 | Onco-mRNA | MRPL42 | 3.48 | 2.47 | 1.41 | 4.10E-07 | 0.013 | 42 | Onco-mRNA | TPI1 | 214.36 | 90.81 | 2.36 | 2.2E-26 | 0.0031 | |||
| 43 | Onco-mRNA | POC1A | 4.59 | 1.41 | 3.27 | 1.6E-32 | 0.00094 | 63 | Onco-mRNA | CBX3 | 47.98 | 20.97 | 2.29 | 1.2E-42 | 0.00063 | |||
| 44 | Onco-mRNA | DARS2 | 10.68 | 4.16 | 2.57 | 2E-33 | 0.00087 | 64 | Onco-mRNA | RPLP0 | 343.61 | 203.06 | 1.69 | 1.3E-11 | 0.037 | |||
| 45 | Onco-mRNA | CDT1 | 5.45 | 0.74 | 7.33 | 1.5E-43 | 0.0019 | 65 | Onco-mRNA | FANCL | 7.35 | 4.66 | 1.58 | 4.8E-13 | 5E-04 | |||
| 46 | Onco-mRNA | KIF14 | 1.56 | 0.11 | 14.04 | 6.6E-57 | 0.00058 | 66 | Onco-mRNA | SKA1 | 2.19 | 0.25 | 8.85 | 1.8E-43 | 0.00043 | |||
| 47 | Onco-mRNA | HMGA1 | 122.27 | 23 | 5.32 | 5.1E-37 | 0.0014 | 67 | Onco-mRNA | FKBP4 | 24.43 | 10.13 | 2.41 | 2E-18 | 2.4E-05 | |||
| 48 | Onco-mRNA | ALDOA | 235.13 | 94.94 | 2.48 | 8.5E-25 | 0.00021 | 68 | Onco-mRNA | AGMAT | 1.55 | 0.24 | 6.39 | 1.4E-49 | 0.0096 | |||
| 49 | Onco-mRNA | CENPI | 1.4 | 0.2 | 7.16 | 1.1E-39 | 0.014 | 69 | Onco-mRNA | CDC25A | 1.44 | 0.26 | 5.52 | 2.6E-38 | 0.014 | |||
| 50 | Onco-mRNA | NOL10 | 9.62 | 5.24 | 1.84 | 1.4E-36 | 0.0054 | 70 | Onco-mRNA | SRM | 35.7 | 15.89 | 2.25 | 1.5E-24 | 0.021 | |||
| 51 | Onco-mRNA | IMP4 | 15.07 | 8.92 | 1.69 | 1.9E-26 | 0.0023 | 71 | Onco-mRNA | OIP5 | 2.34 | 0.42 | 5.64 | 1.9E-35 | 5.7E-05 | |||
| 52 | Onco-mRNA | TMCO1 | 24.58 | 13.89 | 1.77 | 9E-24 | 0.019 | 72 | TS-mRNA | NFIX | 13.42 | 33.09 | 0.41 | 5.9E-25 | 0.007 | |||
| 53 | Onco-mRNA | TMEM106B | 9.24 | 4.54 | 2.03 | 1.3E-15 | 0.0051 | 73 | TS-mRNA | METTL7A | 19.34 | 64.98 | 0.3 | 3.7E-41 | 0.0067 | |||
| 54 | Onco-mRNA | KIF11 | 5.96 | 0.84 | 7.11 | 4.5E-52 | 0.00048 | 74 | TS-mRNA | BTG2 | 59.21 | 131.34 | 0.45 | 1.4E-17 | 0.0014 | |||
| 55 | Onco-mRNA | MTHFD2 | 18.74 | 5.92 | 3.17 | 6.2E-29 | 0.0038 | 75 | TS-mRNA | SNX20 | 2.26 | 2.99 | 0.76 | 0.0000055 | 0.012 | |||
| 56 | Onco-mRNA | TWF1 | 18.66 | 8.99 | 2.08 | 9.5E-23 | 0.00014 | 76 | TS-mRNA | PIK3R1 | 5.03 | 9.26 | 0.54 | 2.8E-20 | 0.00053 | |||
| 57 | Onco-mRNA | YKT6 | 35.3 | 18.72 | 1.89 | 5E-22 | 0.017 | 77 | TS-mRNA | LIFR | 6.34 | 17.36 | 0.37 | 1.9E-25 | 0.035 | |||
| 58 | Onco-mRNA | GTF2E2 | 12.55 | 7.42 | 1.69 | 9.8E-20 | 0.0086 | 78 | TS-mRNA | ABCB1 | 0.73 | 1.92 | 0.38 | 5E-18 | 0.013 | |||
| 59 | Onco-mRNA | SFXN1 | 6.8 | 2.19 | 3.11 | 7.2E-67 | 0.042 | 79 | TS-mRNA | SNX30 | 6.97 | 12.69 | 0.55 | 2E-15 | 0.00091 | |||
| 60 | Onco-mRNA | GCLC | 14.69 | 2.82 | 5.21 | 2.9E-09 | 0.024 | 80 | TS-mRNA | DOCK4 | 2.87 | 7.44 | 0.39 | 8.1E-36 | 0.026 | |||
| 61 | Onco-mRNA | KPNB1 | 37.6 | 21.33 | 1.76 | 8.6E-24 | 0.0027 | 81 | TS-mRNA | GPD1L | 14.94 | 23.51 | 0.64 | 5.9E-13 | 0.0036 | |||
| 62 | Onco-mRNA | TMED2 | 108.86 | 57.26 | 1.9 | 5E-25 | 0.002 | 82 | TS-mRNA | SCD5 | 4.44 | 11.89 | 0.37 | 4.2E-24 | 0.035 | |||
Table 3
Expression correlations of miRNA-mRNA pairs"
| No. | miRNA | gene | Correlation | No. | miRNA | gene | Correlation | No. | miRNA | gene | Correlation | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | P value | R | P value | R | P value | |||||||||||
| 1 | hsa-let-7c-5p | ESPL1 | -0.431 | 1.25E-24 | 34 | hsa-mir-140-3p | CDCA8 | -0.268 | 7.27E-10 | 67 | hsa-mir-30d-5p | MTHFD2 | -0.401 | 3.74E-21 | ||
| 2 | hsa-let-7c-5p | CCNB2 | -0.384 | 2.04E-19 | 35 | hsa-mir-140-3p | CDC25A | -0.203 | 3.79E-06 | 68 | hsa-mir-30d-5p | TWF1 | -0.4 | 4.84E-21 | ||
| 3 | hsa-let-7c-5p | RRM2 | -0.384 | 1.81E-19 | 36 | hsa-mir-140-3p | DNAJC9 | -0.171 | 0.000106 | 69 | hsa-mir-30d-5p | YKT6 | -0.344 | 1.21E-15 | ||
| 4 | hsa-let-7c-5p | CDC25A | -0.361 | 3.13E-17 | 37 | hsa-mir-140-3p | UCK2 | -0.103 | 0.0202 | 70 | hsa-mir-30d-5p | GTF2E2 | -0.329 | 2.20E-14 | ||
| 5 | hsa-let-7c-5p | HMGA1 | -0.348 | 5.20E-16 | 38 | hsa-mir-150-5p | SMUG1 | -0.264 | 1.23E-09 | 71 | hsa-mir-30d-5p | SFXN1 | -0.301 | 3.59E-12 | ||
| 6 | hsa-let-7c-5p | FANCI | -0.332 | 1.17E-14 | 39 | hsa-mir-150-5p | PDIA6 | -0.251 | 8.44E-09 | 72 | hsa-mir-30d-5p | GCLC | -0.245 | 1.91E-08 | ||
| 7 | hsa-let-7c-5p | YWHAZ | -0.296 | 7.73E-12 | 40 | hsa-mir-150-5p | CACYBP | -0.245 | 2.00E-08 | 73 | hsa-mir-30d-5p | KPNB1 | -0.245 | 1.95E-08 | ||
| 8 | hsa-let-7c-5p | GARS | -0.288 | 3.19E-11 | 41 | hsa-mir-150-5p | ALDOA | -0.239 | 4.19E-08 | 74 | hsa-mir-30d-5p | TMED2 | -0.214 | 9.84E-07 | ||
| 9 | hsa-let-7c-5p | BZW1 | -0.282 | 7.79E-11 | 42 | hsa-mir-150-5p | INTS7 | -0.188 | 1.93E-05 | 75 | hsa-mir-30d-5p | CBX3 | -0.19 | 1.53E-05 | ||
| 10 | hsa-let-7c-5p | EIF4A3 | -0.278 | 1.50E-10 | 43 | hsa-mir-150-5p | SLC7A11 | -0.154 | 0.000489 | 76 | hsa-mir-30d-5p | RPLP0 | -0.129 | 0.00346 | ||
| 11 | hsa-let-7c-5p | RHOV | -0.278 | 1.44E-10 | 44 | hsa-mir-150-5p | MRPS10 | -0.153 | 0.000499 | 77 | hsa-mir-30d-5p | FANCL | -0.119 | 0.00722 | ||
| 12 | hsa-let-7c-5p | LDHA | -0.27 | 5.22E-10 | 45 | hsa-mir-150-5p | SLC2A1 | -0.146 | 0.000919 | 78 | hsa-mir-328-3p | RAD51 | -0.188 | 1.76E-05 | ||
| 13 | hsa-let-7c-5p | DIABLO | -0.254 | 5.90E-09 | 46 | hsa-mir-150-5p | BIRC5 | -0.137 | 0.00182 | 79 | hsa-mir-328-3p | SKA1 | -0.141 | 0.00133 | ||
| 14 | hsa-let-7c-5p | MRPL12 | -0.247 | 1.53E-08 | 47 | hsa-mir-150-5p | TTLL12 | -0.133 | 0.00258 | 80 | hsa-mir-328-3p | FKBP4 | -0.1 | 0.0243 | ||
| 15 | hsa-let-7c-5p | POLR2D | -0.225 | 2.52E-07 | 48 | hsa-mir-150-5p | TIMM50 | -0.109 | 0.0139 | 81 | hsa-mir-195-3p | AGMAT | -0.188 | 1.86E-05 | ||
| 16 | hsa-let-7c-5p | CALU | -0.214 | 9.69E-07 | 49 | hsa-mir-1976 | CKS1B | -0.344 | 1.16E-15 | 82 | hsa-mir-195-3p | CDC25A | -0.302 | 2.89E-12 | ||
| 17 | hsa-let-7c-5p | NME4 | -0.181 | 0.0000372 | 50 | hsa-mir-1976 | GPN1 | -0.338 | 4.14E-15 | 83 | hsa-mir-490-3p | SRM | -0.149 | 0.000748 | ||
| 18 | hsa-let-7c-5p | PMAIP1 | -0.169 | 0.000123 | 51 | hsa-mir-1976 | RAD51 | -0.285 | 4.67E-11 | 84 | hsa-mir-490-3p | RAD51 | -0.151 | 0.0006 | ||
| 19 | hsa-let-7c-5p | INTS7 | -0.163 | 0.000218 | 52 | hsa-mir-1976 | TPI1 | -0.28 | 1.10E-10 | 85 | hsa-mir-526b-5p | OIP5 | 0.114 | 0.00992 | ||
| 20 | hsa-let-7c-5p | RFC2 | -0.145 | 0.000963 | 53 | hsa-mir-1976 | POC1A | -0.261 | 2.02E-09 | 86 | hsa-mir-1293 | NFIX | -0.235 | 7.15E-08 | ||
| 21 | hsa-let-7c-5p | IGF2BP3 | -0.14 | 0.00147 | 54 | hsa-mir-1976 | DARS2 | -0.235 | 7.65E-08 | 87 | hsa-mir-873-5p | METTL7A | -0.19 | 1.56E-05 | ||
| 22 | hsa-mir-101-3p | RRM2 | -0.41 | 3.49E-22 | 55 | hsa-mir-1976 | CDT1 | -0.215 | 9.35E-07 | 88 | hsa-mir-873-5p | BTG2 | -0.173 | 8.56E-05 | ||
| 23 | hsa-mir-101-3p | KPNA2 | -0.388 | 7.83E-20 | 56 | hsa-mir-1976 | KIF14 | -0.199 | 5.61E-06 | 89 | hsa-mir-550a-5p | SNX20 | -0.153 | 0.000531 | ||
| 24 | hsa-mir-101-3p | KIF2C | -0.373 | 2.46E-18 | 57 | hsa-mir-1976 | HMGA1 | -0.198 | 6.42E-06 | 90 | hsa-mir-21-5p | BTG2 | -0.301 | 3.61E-12 | ||
| 25 | hsa-mir-101-3p | BIRC5 | -0.301 | 3.66E-12 | 58 | hsa-mir-1976 | ALDOA | -0.187 | 2.06E-05 | 91 | hsa-mir-21-5p | PIK3R1 | -0.218 | 6.69E-07 | ||
| 26 | hsa-mir-101-3p | BZW1 | -0.261 | 2.12E-09 | 59 | hsa-mir-1976 | CENPI | -0.185 | 2.55E-05 | 92 | hsa-mir-21-5p | LIFR | -0.198 | 6.29E-06 | ||
| 27 | hsa-mir-101-3p | MRPL42 | -0.22 | 5.27E-07 | 60 | hsa-mir-1976 | NOL10 | -0.18 | 4.16E-05 | 93 | hsa-mir-21-5p | ABCB1 | -0.185 | 2.44E-05 | ||
| 28 | hsa-mir-101-3p | GCLC | -0.216 | 7.94E-07 | 61 | hsa-mir-1976 | IMP4 | -0.173 | 8.01E-05 | 94 | hsa-mir-21-5p | SNX30 | -0.172 | 8.75E-05 | ||
| 29 | hsa-mir-101-3p | DDIT4 | -0.1 | 0.0237 | 62 | hsa-mir-1976 | RFC2 | -0.168 | 0.00013 | 95 | hsa-mir-21-5p | DOCK4 | -0.137 | 0.00184 | ||
| 30 | hsa-mir-140-3p | TPI1 | -0.403 | 1.99E-21 | 63 | hsa-mir-1976 | TMCO1 | -0.16 | 0.000273 | 96 | hsa-mir-21-5p | GPD1L | -0.133 | 0.00265 | ||
| 31 | hsa-mir-140-3p | AHCY | -0.341 | 2.01E-15 | 64 | hsa-mir-1976 | TMEM106B | -0.108 | 0.0141 | 97 | hsa-mir-9-5p | SCD5 | -0.1 | 0.023 | ||
| 32 | hsa-mir-140-3p | RACGAP1 | -0.296 | 8.31E-12 | 65 | hsa-mir-30d-5p | RRM2 | -0.535 | 2.68E-39 | |||||||
| 33 | hsa-mir-140-3p | CDCA4 | -0.269 | 5.98E-10 | 66 | hsa-mir-30d-5p | KIF11 | -0.416 | 7.76E-23 | |||||||

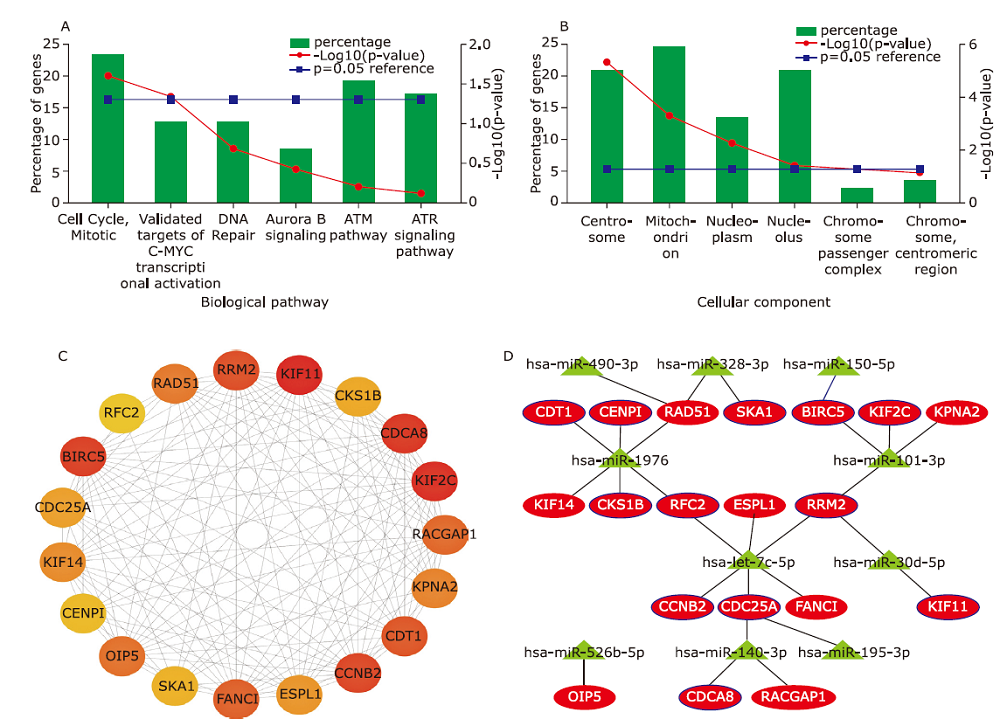
Figure 2.
The miRNA-mRNA regulatory network associated with prognosis of LUAD. miRNAs are represented by triangles; mRNAs are represented by circles; all up-regulated RNAs are in red; all down-regulated RNAs are in green; experimentally validated interactions (miRTarBase) of RNA pairs are marked by bold blue lines."

Table 4
Enriched pathways of key up-regulated mRNAs"
| Biological pathway | Genes mapped (from input data set) |
|---|---|
| Cell Cycle, Mitotic | CCNB2; RRM2; CDC25A; RFC2; KIF2C; BIRC5; CDCA8; CKS1B; CDT1; CENPI; SKA1 |
| Validated targets of C-MYC transcriptional activation | CDC25A; HMGA1; LDHA; PMAIP1; BIRC5; SLC2A1 |
Table 5
qPCR primers"
| Primer name | sequence (5’ to 3’) | Primer name | sequence (5’ to 3’) | |
|---|---|---|---|---|
| Q-H-CDT1-F | TGTCAAGGAGCACCACAAGGC | Q-H-BIRC5-F | TCTCAAGGACCACCGCATCT | |
| Q-H-CDT1-R | CGATGTCGGGTACTTCATCCA | Q-H-BIRC5-R | CCAAGTCTGGCTCGTTCTCA | |
| Q-H-CENPI-F | CTCAGATGGGCTCAGTGCTAA | Q-H-KIF2C-F | AGAATTTCGGGCTACTTTGG | |
| Q-H-CENPI-R | TGAAGGAACTGTGACCGACAA | Q-H-KIF2C-R | TGCTTATTCAGTGGGCGTTT | |
| Q-H-RAD51-F | TGTCAAGGAGCACCACAAGGC | Q-H-KPNA2-F | CGTCGCAGAATAGAGGTCAA | |
| Q-H-RAD51-R | CGATGTCGGGTACTTCATCCA | Q-H-KPNA2-R | GCAGCGGAGAAGTAGCATCA | |
| Q-H-KIF14-F | AAAAGAAAGTCTGGGTGGA | Q-H-CCNB2-F | CGAGGACATTGATAACGAAG | |
| Q-H-KIF14-R | CTGTATCGTTCAGGGTCAA | Q-H-CCNB2-R | AACTGGCTGAACCTGTAAAA | |
| Q-H-CKS1B-F | TCATGCTGCCCAAGGACATA | Q-H-CDC25A-F | AGGGTCTGGGCAGTGATTAT | |
| Q-H-CKS1B-R | GGACCCATCCCTGACTCTGC | Q-H-CDC25A-R | AACAGCTTCTGAGGTAGGGA | |
| Q-H-RFC2-F | CTTCTCAGGATTTGGCTTCA | Q-H-FANCI-F | GCTCTGAGGAAGCTGGAACA | |
| Q-H-RFC2-R | GTAGGCTTCGTCAATGTTGG | Q-H-FANCI-R | AGGGCAGTGAGAATGATAGG | |
| Q-H-ESPL1-F | GCTTGTGATGCCATCCTGAGGG | Q-H-CDCA8-F | TGAGTCAGACAGGCAGAACC | |
| Q-H-ESPL1-R | GCCTCTGGGCTTCCTTGTGC | Q-H-CDCA8-R | TTCCTCCAAGGGCGAAGTAG | |
| Q-H-RRM2-F | AACTTGGTGGAGCGATTTAG | Q-H-RACGAP1-F | AGCGAAGTGCTCTGGATGTT | |
| Q-H-RRM2-R | ATAGGTAGCCTCTTTGTCCC | Q-H-RACGAP1-R | CATTGCTGCTGGATGGTTGG | |
| Q-H-ACTB-F | ACTCTTCCAGCCTTCCTTCC | |||
| Q-H-ACTB-R | TGTTGGCGTACAGGTCTTTG |
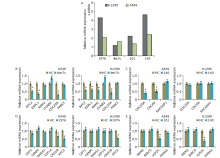
Figure 4.
Experimental validation of miRNA-hub gene pairs. Relative miRNA expression (to blank control) after transfected with1976, 101, let-7c,or140 in A549 and H1299; relative mRNA expression of predicted targets after over-expression of 1976 (A), 101 (B), let-7c (C), and 140 (D) in A549 and H1299. NC: natural control mimic; 1976: hsa-miR-1976 mimic; 101: hsa-miR-101-3p mimic; let-7c: hsa-let-7c-5p mimic; 140: hsa-miR-140-3pmimic."
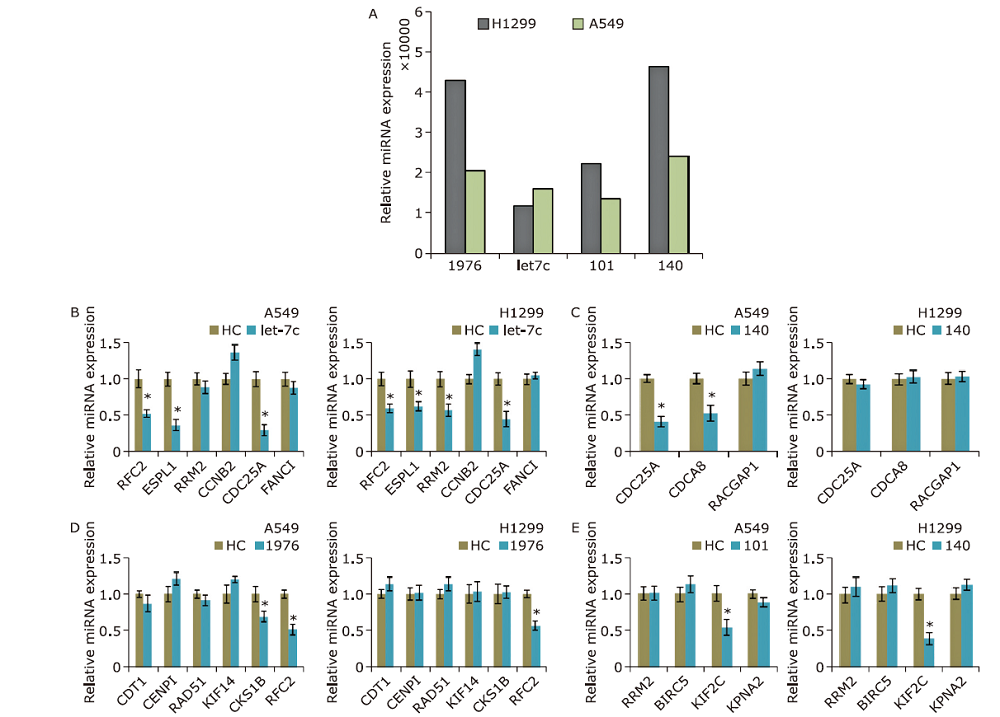

Figure 5.
Influences of miRNA over-expression on cell cycle. hsa-miR-1976 and hsa-let-7c-5p over-expression induced the G0/G1 phase arrest of A549 cells. NC: natural control mimic; 1976: hsa-miR-1976 mimic; 101: hsa-miR-101-3p mimic; let-7c: hsa-let-7c-5p mimic; 140: hsa-miR-140-3p mimic."

| 1. |
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69(1):7-34. doi: 10.3322/caac.21551.
doi: 10.3322/caac.21551 |
| 2. |
Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016; 893:1-19. doi: 10.1007/978-3-319-24223-1_1.
doi: 10.1007/978-3-319-24223-1_1 |
| 3. |
He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447(7148):1130-4. doi: 10.1038/nature05939.
doi: 10.1038/nature05939 |
| 4. |
Misso G, Di Martino MT, De Rosa G, et al. miR-34: a new weapon against cancer?. Mol Ther Nucleic Acids 2014; 3(9):e194. doi: 10.1038/mtna.2014.47.
doi: 10.1038/mtna.2014.47 |
| 5. |
Shi Y, Liu C, Liu X, et al. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One 2014; 9(3):e90022. doi: 10.1371/journal.pone.0090022.
doi: 10.1371/journal.pone.0090022 |
| 6. |
Cortez MA, Ivan C, Valdecanas D, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst 2016; 108(1): djv303. doi: 10.1093/jnci/djv303.
doi: 10.1093/jnci/djv303 |
| 7. |
Ceppi P, Mudduluru G, Kumarswamy R, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res 2010; 8(9):1207-16. doi: 10.1158/1541-7786.MCR-10-0052.
doi: 10.1158/1541-7786.MCR-10-0052 |
| 8. |
Takeyama Y, Sato M, Horio M, et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett 2010; 296(2):216-24. doi: 10.1016/j.canlet.2010.04.008.
doi: 10.1016/j.canlet.2010.04.008 |
| 9. |
Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009; 23(18):2140-51. doi: 10.1101/gad.1820209.
doi: 10.1101/gad.1820209 |
| 10. |
Cortez MA, Valdecanas D, Zhang X, et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol Ther 2014; 22(8):1494-503. doi: 10.1038/mt.2014.79.
doi: 10.1038/mt.2014.79 |
| 11. |
Wang X, Ding Y, Da B, et al. Identification of potential prognostic long noncoding RNA signatures based on a competing endogenous RNA network in lung adenocarcinoma. Oncol Rep 2018; 40(6):3199-212. doi: 10.3892/or.2018.6719.
doi: 10.3892/or.2018.6719 |
| 12. |
Wang XW, Guo QQ, Wei Y, et al. Construction of a competing endogenous RNA network using differentially expressed lncRNAs, miRNAs and mRNAs in nonsmall cell lung cancer. Oncol Rep 2019; 42(6):2402-15. doi: 10.3892/or.2019.7378.
doi: 10.3892/or.2019.7378 |
| 13. |
Wong NW, Chen Y, Chen S, et al. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018; 34(4):713-5. doi: 10.1093/bioinformatics/btx627.
doi: 10.1093/bioinformatics/btx627 |
| 14. |
Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42(Database issue):D92-7. doi: 10.1093/nar/gkt1248.
doi: 10.1093/nar/gkt1248 |
| 15. |
Chang L, Zhou G, Soufan O, et al. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res 2020; 48(W1):W244-W251. doi: 10.1093/nar/gkaa467.
doi: 10.1093/nar/gkaa467 |
| 16. |
Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017; 19(8):649-58. doi: 10.1016/j.neo.2017.05.002.
doi: S1476-5586(17)30179-3 pmid: 28732212 |
| 17. |
Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018; 46(D1):D296-D302. doi: 10.1093/nar/gkx1067.
doi: 10.1093/nar/gkx1067 |
| 18. |
Pathan M, Keerthikumar S, Ang CS, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015; 15(15):2597-601. doi: 10.1002/pmic.201400515.
doi: 10.1002/pmic.201400515 |
| 19. |
Szklarczyk D, Morris J H, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45(D1):D362-D368. doi: 10.1093/nar/gkw937.
doi: 10.1093/nar/gkw937 |
| 20. |
Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int 2015; 15:38. doi: 10.1186/s12935-015-0185-1.
doi: 10.1186/s12935-015-0185-1 |
| 21. |
Fang L, Cai J, Chen B, et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/beta-catenin signalling. Nat Commun 2015; 6:8640. doi: 10.1038/ncomms9640.
doi: 10.1038/ncomms9640 |
| 22. |
Luo J, Zhu H, Jiang H, et al. The effects of aberrant expression of LncRNA DGCR5/miR-873-5p/TUSC3 in lung cancer cell progression. Cancer Med 2018; 7(7):3331-41. doi: 10.1002/cam4.1566.
doi: 10.1002/cam4.1566 |
| 23. | Hosseini SM, Soltani BM, Tavallaei M, et al. Clinically Significant Dysregulation of hsa-miR-30d-5p and hsa-let-7b Expression in Patients with Surgically Resected Non-Small Cell Lung Cancer. Avicenna J Med Biotechnol 2018; 10(2):98-104. |
| 24. |
Wang P, Zou F, Zhang X, et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 2009; 69(20):8157-65. doi: 10.1158/0008-5472.CAN-09-1996.
doi: 10.1158/0008-5472.CAN-09-1996 |
| 25. |
Hu T, Shen H, Li J, et al. RFC2, a direct target of miR-744, modulates the cell cycle and promotes the proliferation of CRC cells. J Cell Physiol 2020; 235(11):8319-33. doi: 10.1002/jcp.29676.
doi: 10.1002/jcp.29676 |
| 26. |
Gan H, Lin L, Hu N, et al. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem Funct 2019; 37(6):424-31. doi: 10.1002/cbf.3420.
doi: 10.1002/cbf.3420 |
| 27. | Huang D, Zheng Z, Huang J, et al. Diagnostic signifcance of circulating let-7c-5p and its potential target gene network in non-small cell lung cancers. Int J Clin Exp Med 2016; 9:9828-42. |
| 28. |
Chen G, Hu J, Huang Z, et al. MicroRNA-1976 functions as a tumor suppressor and serves as a prognostic indicator in non-small cell lung cancer by directly targeting PLCE1. Biochem Biophys Res Commun 2016; 473(4):1144-1151. doi: 10.1016/j.bbrc.2016.04.030.
doi: 10.1016/j.bbrc.2016.04.030 |
| [1] | Li Wenxing, Zhang Yanli. Novel Long Non-coding RNA Markers for Prognostic Prediction of Patients with Bladder Cancer [J]. Chinese Medical Sciences Journal, 2020, 35(3): 239-247. |
| [2] | Zhu Weihua,Xie Wenyong,Zhang Zhedong,Li Shu,Zhang Dafang,Liu Yijun,Zhu Jiye,Leng Xisheng. Postoperative Complications and Survival Analysis of Surgical Resection for Hilar Cholangiocarcinoma: A Retrospective Study of Fifty-Nine Consecutive Patients [J]. Chinese Medical Sciences Journal, 2020, 35(2): 157-169. |
| [3] | Chen Qiang, Zhang Liwei, Huang Dangsheng, Zhang Chunhong, Wang Qiushuang, Shen Dong, Xiong Minjun, Yang Feifei. Five-year Clinical Outcomes of CAD Patients Complicated with Diabetes after StentBoost-optimized Percutaneous Coronary Intervention [J]. Chinese Medical Sciences Journal, 2019, 34(3): 177-183. |
| [4] | Wang Guorong, Wang Zhiwei, Jin Zhengyu. Application and Progress of Texture Analysis in the Therapeutic Effect Prediction and Prognosis of Neoadjuvant Chemoradiotherapy for Colorectal Cancer [J]. Chinese Medical Sciences Journal, 2019, 34(1): 45-50. |
| [5] | Liu Yongsheng, Zhao Yu. Progress in Intraoperative Neurophysiological Monitoring for the Surgical Treatment of Thoracic Spinal Stenosis [J]. Chinese Medical Sciences Journal, 2017, 32(4): 260-264. |
| [6] | Meng-yi Wang, Zhe-yu Niu, Xiang-Gao, Li Zhou, Quan Liao, Yu-pei Zhao. Prognostic Impact of Cell Division Cycle Associated 2 Expression on Pancreatic Ductal Adenocarcinoma [J]. Chinese Medical Sciences Journal, 2016, 31(3): 149-154. |
| [7] | Min Xu, Zheng-song Gu, Cun-zu Wang, Xiao-feng Lu, Ding-chao Xiang, Zhi-cheng Yuan, Qiao-yu Li, Min Wu. Impact of Intraoperative Blood Pressure Control and Temporary Parent Artery Blocking on Prognosis in Cerebral Aneurysms Surgery [J]. Chinese Medical Sciences Journal, 2016, 31(2): 89-94. |
| [8] | Shu-bo Tian, Jian-chun Yu*, Wei-ming Kang, Zhi-qiang Ma, Xin Ye, Chao Yan, Ya-kai Huang. Effect of Neoadjuvant Chemotherapy Treatment on Prognosis of Patients with Advanced Gastric Cancer: a Retrospective Study [J]. Chinese Medical Sciences Journal, 2015, 30(2): 84-89. |
| [9] | Ying-qiu Pan, Wei-wu Shi, Dan-ping Xu, Hui-hui Xu, Mei-ying Zhou, Wei-hua Yan*. Associations Between Epidermal Growth Factor Receptor Gene Mutation and Serum Tumor Markers in Advanced Lung Adenocarcinomas: A Retrospective Study [J]. Chinese Medical Sciences Journal, 2014, 29(3): 156-161. |
| [10] | Xiao-dong Qu, Resha Shrestha and Mao-de Wang* . Risk Factors Analysis on Traumatic Brain Injury Prognosis [J]. Chinese Medical Sciences Journal, 2011, 26(2): 98-102. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
|