Chinese Medical Sciences Journal ›› 2023, Vol. 38 ›› Issue (3): 191-205.doi: 10.24920/004223
• Original Article • Previous Articles Next Articles
Cuproptosis-Related 4-Gene Risk Model for Predicting Immunotherapy Drug Response and Prognosis of Kidney Renal Clear Cell Carcinoma
Jin-Shuai Guo1, Hao Ding1, Peng-Yu Wu1, Zi-Yi Xin1, Jian-Xin Li1, Hyon-Su Jo2, Zhen-Hai Ma( )
)
- 1Department of Breast Surgery, Breast Cancer Key Lab of Dalian, the Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning 116027, China
2Department of General Surgery, the Hospital of Pyongyang Medical University, D.P.R. Korea
-
Received:2023-03-16Accepted:2023-06-12Published:2023-09-30Online:2023-07-28 -
Contact:* Email:mazhenhai@dmu.edu.cn .
| Kidney renal clear cell carcinoma (KIRC) is the most common pathological type of renal cell carcinoma. This study adds to the understanding of the molecular mechanisms of KIRC and demonstrates the potential relationship between cuproptosis, the immune microenvironment, and the prognosis of KIRC patients based on bioinformatics. |
Cite this article
Jin-Shuai Guo, Hao Ding, Peng-Yu Wu, Zi-Yi Xin, Jian-Xin Li, Hyon-Su Jo, Zhen-Hai Ma. Cuproptosis-Related 4-Gene Risk Model for Predicting Immunotherapy Drug Response and Prognosis of Kidney Renal Clear Cell Carcinoma[J].Chinese Medical Sciences Journal, 2023, 38(3): 191-205.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1.
Clinical characteristics of the KIRC patients"
| TCGA-KIRC dataset [n(%)] | GEO-GSE29609 [n(%)] | ||
|---|---|---|---|
| Age | < 60y | 247 (46.0%) | 15 (38.5%) |
| ≥60y | 290 (54.0%) | 24 (61.5%) | |
| Gender | Female | 191 (35.6%) | - |
| Male | 346 (64.4%) | - | |
| Grade | Grade 1 | 14 (2.6%) | 1 (2.6%) |
| Grade 2 | 230 (42.8%) | 12 (30.8%) | |
| Grade 3 | 207 (38.6%) | 11 (28.2%) | |
| Grade 4 | 78 (14.5%) | 15 (39.4%) | |
| unknown | 8 (1.5%) | 0 (0.0%) | |
| Stage | Stage Ⅰ | 269 (50.1%) | - |
| Stage Ⅱ | 57 (10.6%) | - | |
| Stage Ⅲ | 125 (23.3%) | - | |
| Stage Ⅳ | 83 (15.4%) | - | |
| unknown | 3 (0.6%) | - | |
| T | T1 | 275 (51.2%) | 11 (28.2%) |
| T2 | 69 (12.9%) | 5 (12.8%) | |
| T3 | 182 (33.9%) | 22 (56.4%) | |
| T4 | 11 (2.0%) | 1 (2.6%) | |
| M | M0 | 426 (79.3%) | 25 (64.1%) |
| M1 | 79 (14.7%) | 14 (35.9%) | |
| unknown | 32 (6.0%) | 0 (0.0%) | |
| N | N0 | 240 (44.7%) | 31 (79.5%) |
| N1 | 17 (3.2%) | 8 (20.5%) | |
| unknown | 280 (52.1%) | 0 (0.0%) |
Figure 2.
Identification of the CRGs in the TCGA dataset. (A) Venn diagram to identify differentially expressed genes. (B) Heatmap to show the expression of genes in Heatmap showing the expression of genes in different groups. Red means the gene is highly expressed, whereas blue means the gene is low expressed. (C) Forest plot to show the results of the univariate Cox regression analysis between gene expression and OS. (D) The PPI network to show the interaction among eight CEGs. (E) The correlation network of eight CEGs The correlation network of eight CEGs. The correlation coefficients are represented by different colors."
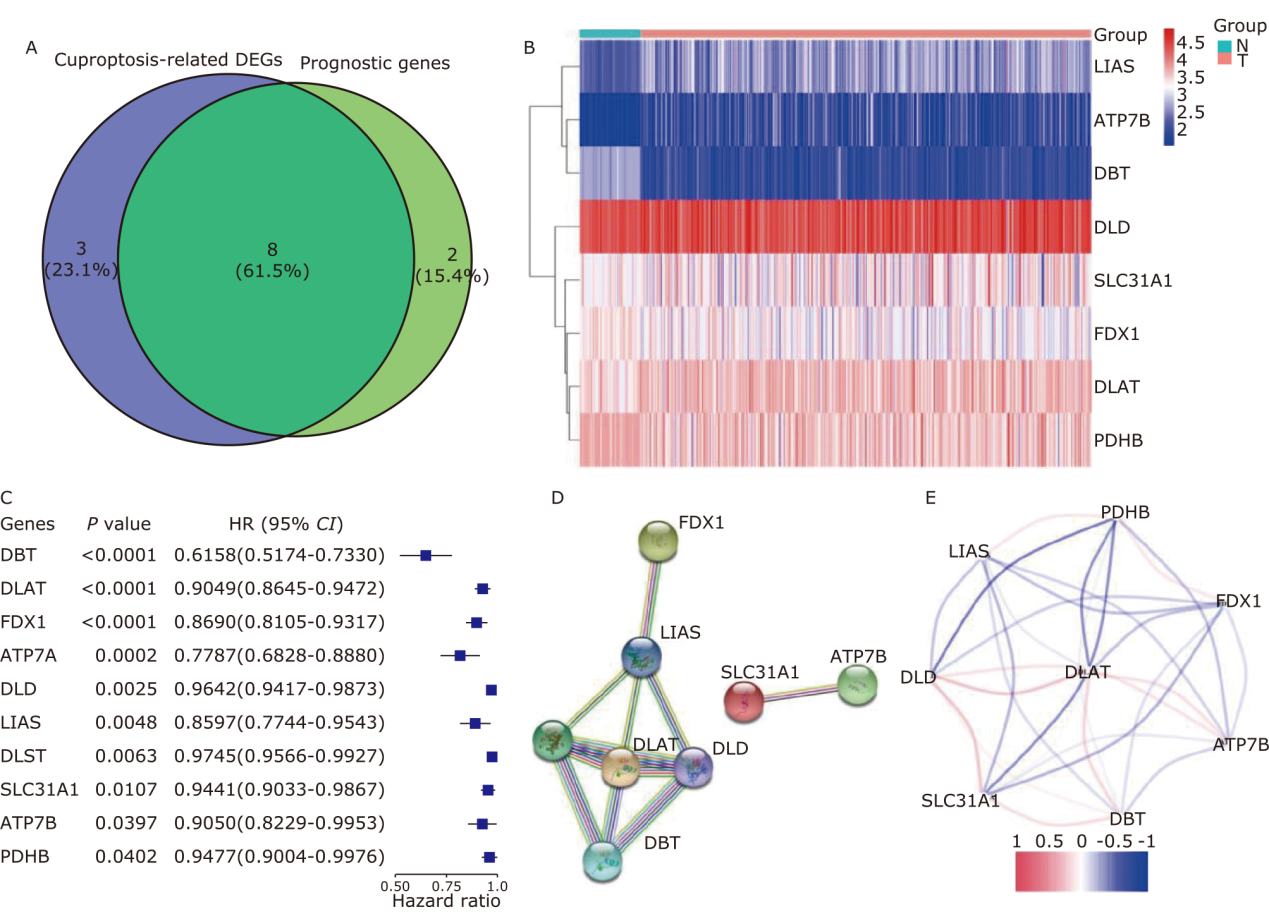

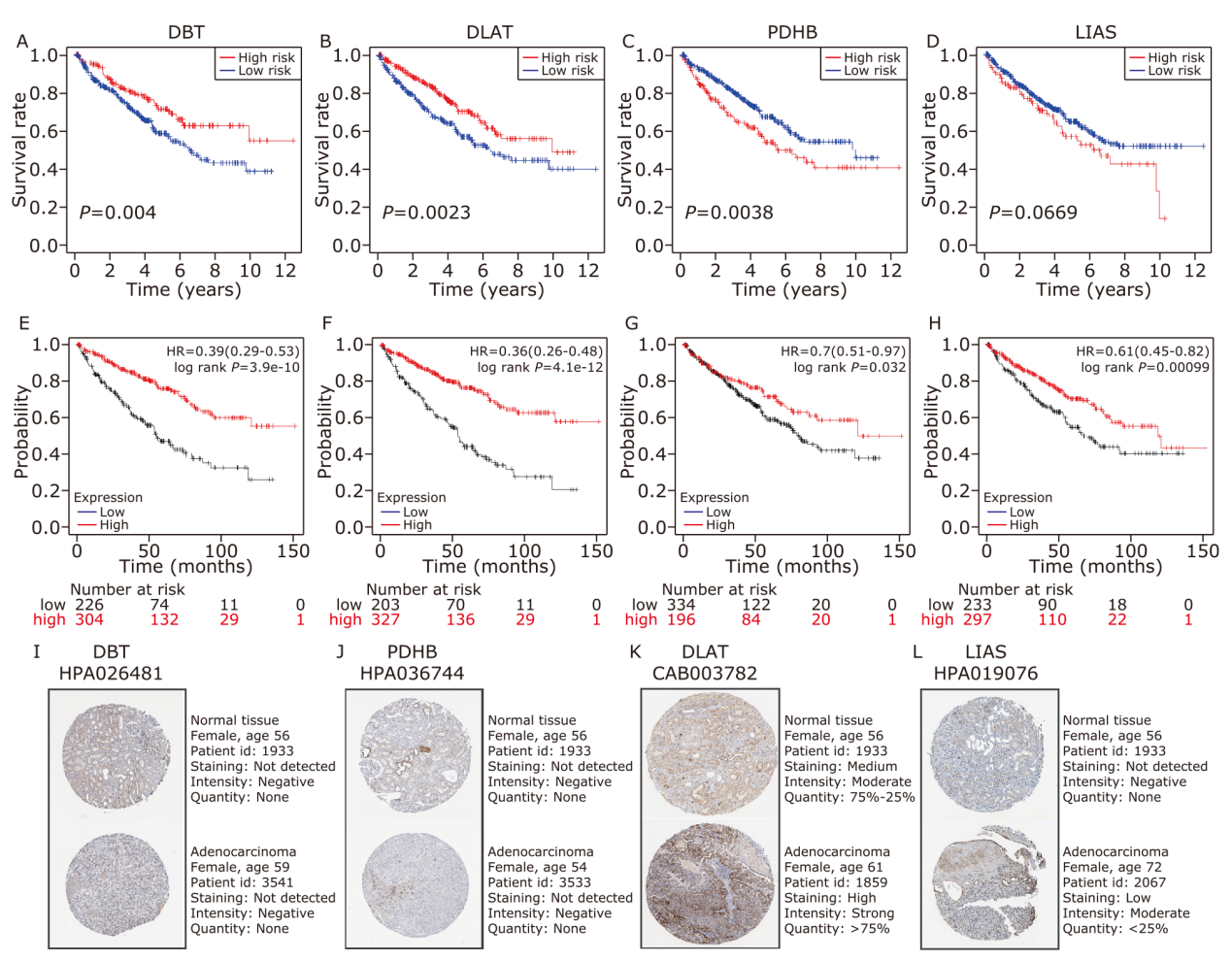
Figure 3.
Prognostic analysis of the 4 CRGs signature model in the TCGA dataset. (A-C) LASSO-Cox regression analysis to identify hub genes. (D) The distribution and median value of the risk scores. (E) PCA plot to show the similarity between the individual samples. (F) The t-SNE analysis to verify the effectiveness of the algorithm. (G) The distributions of OS status, OS and risk score in the TCGA dataset. (H) KM curves for the OS of patients in the high- and low-risk groups. (I) The AUC of the time-dependent ROC curves to verify the prognostic performance of the risk score."

Table 2.
Baseline characteristics of the patients in different risk groups"
| Characteristics | TCGA-KIRC dataset | GEO-GSE29609 dataset | |||||
|---|---|---|---|---|---|---|---|
| High risk | Low risk | P value | High risk | Low risk | P value | ||
| Age [n (%)] | 0.127 | 0.595 | |||||
| <60y | 56 (47.5) | 47 (37.0) | 6 (31.6) | 9 (45.0) | |||
| ≥60y | 62 (52.5) | 80 (63.0) | 13 (68.4) | 11 (55.0) | |||
| Gender [n (%)] | 0.328 | - | |||||
| Female | 42 (35.6) | 54 (42.5) | - | - | |||
| Male | 76 (64.4) | 73 (57.5) | - | - | |||
| Grade [n (%)] | 0.093 | 1 | |||||
| G1+G2 | 45 (38.1) | 63 (49.6) | 6 (31.6) | 7 (35.0) | |||
| G3+G4 | 73 (61.9) | 64 (50.4) | 13 (68.4) | 13 (65.0) | |||
| Stage [n (%)] | 0.038 | - | |||||
| Ⅰ+Ⅱ | 55 (46.7) | 77 (60.6) | - | - | |||
| Ⅲ+Ⅳ | 63 (53.3) | 50 (39.4) | - | - | |||
| T [n (%)] | 0.207 | 0.399 | |||||
| T1+T2 | 64 (54.2) | 80 (63.0) | 6 (31.6) | 10 (50.0) | |||
| T3+T4 | 54 (45.8) | 47 (37.0) | 13 (68.4) | 10 (50.0) | |||
| N [n (%)] | 0.677 | 0.633 | |||||
| N0 | 110 (93.2) | 121 (95.3) | 14 (73.7) | 17 (85.0) | |||
| N1 | 8 (6.8) | 6 (4.7) | 5 (26.3) | 3 (15.0) | |||
| M [n (%)] | 0.008 | 0.121 | |||||
| M0 | 90 (76.3) | 114 (89.8) | 15 (78.9) | 10 (50.0) | |||
| M1 | 28 (23.7) | 13 (10.2) | 4 (21.1) | 10 (50.0) | |||
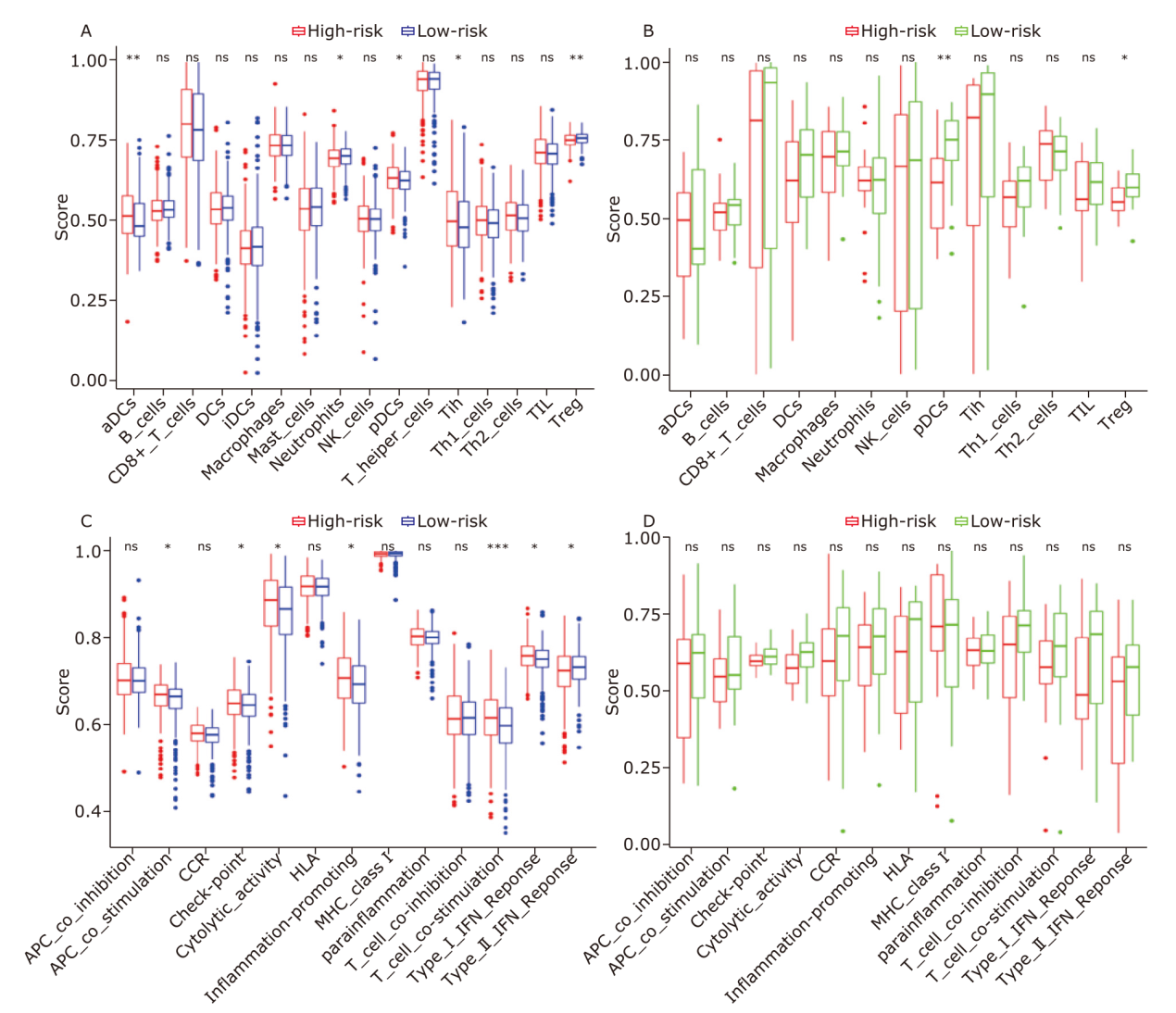
Figure 6.
Comparison of the ssGSEA scores between different risk groups. The boxplots showing 16 immune cells (A, B) and 13 immune-related functions (C, D) between different risk groups in the TCGA dataset (A, C) and the GEO dataset (B, D). P values are showed as: ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001."
| 1 | Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022; 72(1): 7-33. doi: 10.3322/caac.21708. |
| 2 |
Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol 2015; 67(5): 913-24. doi: 10.1016/j.eururo.2015.01.005.
pmid: 25616710 |
| 3 |
Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009; 373(9669): 1119-32. doi: 10.1016/s0140-6736(09)60229-4.
pmid: 19269025 |
| 4 |
Zhang C, Chen L, Liu Y, et al. Downregulated METTL14 accumulates BPTF that reinforces super-enhancers and distal lung metastasis via glycolytic reprogramming in renal cell carcinoma. Theranostics 2021; 11(8): 3676-93. doi: 10.7150/thno.55424.
pmid: 33664855 |
| 5 | Colbert LE, Fisher SB, Balci S, et al. High nuclear hypoxia-inducible factor 1 alpha expression is a predictor of distant recurrence in patients with resected pancreatic adenocarcinoma. Int J Radiat Oncol Biol Phys 2015; 91(3): 631-9. doi: 10.1016/j.ijrobp.2014.11.004. |
| 6 |
Elfiky AA, Aziz SA, Conrad PJ, et al. Characterization and targeting of phosphatidylinositol-3 kinase (PI3K) and mammalian target of rapamycin (mTOR) in renal cell cancer. J Transl Med 2011; 9: 133. doi: 10.1186/1479-5876-9-133.
pmid: 21834980 |
| 7 |
Chen L, Luo Y, Wang G, et al. Prognostic value of a gene signature in clear cell renal cell carcinoma. J Cell Physiol 2019; 234(7): 10324-35. doi: 10.1002/jcp.27700.
pmid: 30417359 |
| 8 | Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol 2008; 4(3): 176-85. doi: 10.1038/nchembio.72. |
| 9 |
Tisato F, Marzano C, Porchia M, et al. Copper in diseases and treatments, and copper-based anticancer strategies. Med Res Rev 2010; 30(4): 708-49. doi: 10.1002/med.20174.
pmid: 19626597 |
| 10 |
Caruano-Yzermans AL, Bartnikas TB, Gitlin JD. Mechanisms of the copper-dependent turnover of the copper chaperone for superoxide dismutase. J Biol Chem 2006; 281(19): 13581-87. doi: 10.1074/jbc.M601580200.
pmid: 16531609 |
| 11 |
Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 2020; 17(7): 395-417. doi: 10.1038/s41571-020-0341-y.
pmid: 32203277 |
| 12 | Weinlich R, Oberst A, Beere HM, et al. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol 2017; 18(2): 127-36. doi: 10.1038/nrm.2016.149. |
| 13 |
Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009; 7(2): 99-109. doi: 10.1038/nrmicro2070.
pmid: 19148178 |
| 14 |
Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012; 149(5): 1060-72. doi: 10.1016/j.cell.2012.03.042.
pmid: 22632970 |
| 15 |
Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022; 375(6586): 1254-61. doi: 10.1126/science.abf0529.
pmid: 35298263 |
| 16 | Ge EJ, Bush AI, Casini A, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer 2022; 22(2): 102-13. doi: 10.1038/s41568-021-00417-2. |
| 17 | Brady DC, Crowe MS, Greenberg DN, et al. Copper chelation inhibits BRAF(V600E)-driven melanomagenesis and counters resistance to BRAF(V600E) and MEK1/2 inhibitors. Cancer Res 2017; 77(22): 6240-52. doi: 10.1158/0008-5472.Can-16-1190. |
| 18 | Yip NC, Fombon IS, Liu P, et al. Disulfiram modulated ROS-MAPK and NFκB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br J Cancer 2011; 104(10): 1564-74. doi: 10.1038/bjc.2011.126. |
| 19 | Tsang T, Posimo JM, Gudiel AA, et al. Copper is an essential regulator of the autophagic kinases ULK1/ 2 to drive lung adenocarcinoma. Nat Cell Biol 2020; 22(4): 412-24. doi: 10.1038/s41556-020-0481-4. |
| 20 | Xiong K, Zhou Y, Karges J, et al. Autophagy-dependent apoptosis induced by apoferritin-Cu(II) nanoparticles in multidrug-resistant colon cancer cells. ACS Appl Mater Interfaces 2021; 13(33): 38959-68. doi: 10.1021/acsami.1c07223. |
| 21 | Parmar A, Pascali G, Voli F, et al. In vivo [(64)Cu]CuCl(2) PET imaging reveals activity of Dextran-Catechin on tumor copper homeostasis. Theranostics 2018; 8(20): 5645-59. doi: 10.7150/thno.29840. |
| 22 |
Lun X, Wells JC, Grinshtein N, et al. Disulfiram when combined with copper enhances the therapeutic effects of temozolomide for the treatment of glioblastoma. Clin Cancer Res 2016; 22(15): 3860-75. doi: 10.1158/1078-0432.Ccr-15-1798.
pmid: 27006494 |
| 23 |
Bandmann O, Weiss KH, Kaler SG. Wilson’s disease and other neurological copper disorders. Lancet Neurol 2015; 14(1): 103-13. doi: 10.1016/s1474-4422(14)70190-5.
pmid: 25496901 |
| 24 |
Gaggelli E, Kozlowski H, Valensin D, et al. Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis). Chem Rev 2006; 106(6): 1995-2044. doi: 10.1021/cr040410w.
pmid: 16771441 |
| 25 |
Jain S, Cohen J, Ward MM, et al. Tetrathiomolybdate-associated copper depletion decreases circulating endothelial progenitor cells in women with breast cancer at high risk of relapse. Ann Oncol 2013; 24(6): 1491-8. doi: 10.1093/annonc/mds654.
pmid: 23406736 |
| 26 | Cui L, Gouw AM, Lagory EL, et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat Biotechnol 2021; 39(3): 357-67. doi: 10.1038/s41587-020-0707-9. |
| 27 | Hu X, Liao S, Bai H, et al. Long noncoding RNA and predictive model to improve diagnosis of clinically diagnosed pulmonary tuberculosis. J Clin Microbiol 2020; 58(7):e01973-19. doi: 10.1128/jcm.01973-19. |
| 28 | Wu Z, Lu Z, Li L, et al. Identification and validation of ferroptosis-related lncRNA signatures as a novel prognostic model for colon cancer. Front Immunol 2021; 12: 783362. doi: 10.3389/fimmu.2021.783362. |
| 29 | Hong HC, Chuang CH, Huang WC, et al. A panel of eight microRNAs is a good predictive parameter for triple-negative breast cancer relapse. Theranostics 2020; 10(19): 8771-89. doi: 10.7150/thno.46142. |
| 30 | Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43(7): e47. doi: 10.1093/nar/gkv007. |
| 31 |
Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021; 49(D1): D605-d12. doi: 10.1093/nar/gkaa1074.
pmid: 33237311 |
| 32 |
Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997; 16(4): 385-95. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3.
pmid: 9044528 |
| 33 | Yu B, Tao D. Heatmap regression via randomized rounding. IEEE Trans Pattern Anal Mach Intell 2022; 44(11): 8276-89. doi: 10.1109/tpami.2021.3103980. |
| 34 | Pontén F, Jirström K, Uhlen M. The Human Protein Atlas: a tool for pathology. J Pathol 2008; 216(4): 387-93. doi: 10.1002/path.2440. |
| 35 | Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123(3): 725-31. doi: 10.1007/s10549-009-0674-9. |
| 36 |
Nagy Á, Munkácsy G, GyŐRffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021; 11(1): 6047. doi: 10.1038/s41598-021-84787-5.
pmid: 33723286 |
| 37 | Wu T, Hu E, Xu S, et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021; 2(3): 100141. doi: 10.1016/j.xinn.2021.100141. |
| 38 |
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14: 7. doi: 10.1186/1471-2105-14-7.
pmid: 23323831 |
| 39 |
Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018; 24(10): 1550-58. doi: 10.1038/s41591-018-0136-1.
pmid: 30127393 |
| 40 | Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014; 9(9): e107468. doi: 10.1371/journal.pone.0107468. |
| 41 | Brady DC, Crowe MS, Turski ML, et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 2014; 509(7501): 492-6. doi: 10.1038/nature13180. |
| 42 | Liao G, Lv J, Ji A, et al. TLR3 serves as a prognostic biomarker and associates with immune infiltration in the renal clear cell carcinoma microenvironment. J Oncol 2021; 2021: 3336770. doi: 10.1155/2021/3336770. |
| 43 | Sun Z, Jing C, Guo X, et al. Comprehensive analysis of the immune infiltrates of pyroptosis in kidney renal clear cell carcinoma. Front Oncol 2021; 11: 716854. doi: 10.3389/fonc.2021.716854. |
| 44 | Xing XL, Liu Y, Liu J, et al. Comprehensive analysis of ferroptosis- and immune-related signatures to improve the prognosis and diagnosis of kidney renal clear cell carcinoma. Front Immunol 2022; 13: 851312. doi: 10.3389/fimmu.2022.851312. |
| 45 | Ahn SH, Yang HY, Tran GB, et al. Interaction of peroxiredoxin Ⅴ with dihydrolipoamide branched chain transacylase E2 (DBT) in mouse kidney under hypoxia. Proteome Sci 2015; 13: 4. doi: 10.1186/s12953-014-0061-2. |
| 46 | Chuang DT, Chuang JL, Wynn RM. Lessons from genetic disorders of branched-chain amino acid metabolism. J Nutr 2006; 136(1 Suppl): 243s-9s. doi: 10.1093/jn/136.1.243S. |
| 47 |
Friedrich T, Lambert AM, Masino MA, et al. Mutation of zebrafish dihydrolipoamide branched-chain transacylase E 2 results in motor dysfunction and models maple syrup urine disease. Dis Model Mech 2012; 5(2): 248-58. doi: 10.1242/dmm.008383.
pmid: 22046030 |
| 48 |
Huang X, Gan G, Wang X, et al. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy 2019; 15(7): 1258-79. doi: 10.1080/15548627.2019.1580105.
pmid: 30786811 |
| 49 |
Goh WQ, Ow GS, Kuznetsov VA, et al. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. Am J Transl Res 2015; 7(6): 1140-51.
pmid: 26279757 |
| 50 |
Thelen JJ, Muszynski MG, David NR, et al. The dihydrolipoamide S-acetyltransferase subunit of the mitochondrial pyruvate dehydrogenase complex from maize contains a single lipoyl domain. J Biol Chem 1999; 274(31): 21769-75. doi: 10.1074/jbc.274.31.21769.
pmid: 10419491 |
| 51 |
Chen S, Liu X, Peng C, et al. The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab 2021; 33(3): 565-80.e7. doi: 10.1016/j.cmet.2021.02.007.
pmid: 33657393 |
| 52 |
Head RA, Brown RM, Zolkipli Z, et al. Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: dihydrolipoamide acetyltransferase (E2) deficiency. Ann Neurol 2005; 58(2): 234-41. doi: 10.1002/ana.20550.
pmid: 16049940 |
| 53 | Yi X, Kim K, Yuan W, et al. Mice with heterozygous deficiency of lipoic acid synthase have an increased sensitivity to lipopolysaccharide-induced tissue injury. J Leukoc Biol 2009; 85(1): 146-53. doi: 10.1189/jlb.0308161. |
| 54 |
Solmonson A, Deberardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem 2018; 293(20): 7522-30. doi: 10.1074/jbc.TM117.000259.
pmid: 29191830 |
| 55 | Hendricks AL, Wachnowsky C, Fries B, et al. Characterization and reconstitution of human lipoyl synthase (LIAS) supports ISCA2 and ISCU as primary cluster donors and an ordered mechanism of cluster assembly. Int J Mol Sci 2021; 22(4). doi: 10.3390/ijms22041598. |
| 56 | Habarou F, Hamel Y, Haack TB, et al. Biallelic mutations in LIPT 2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am J Hum Genet 2017; 101(2): 283-90. doi: 10.1016/j.ajhg.2017.07.001. |
| 57 |
Mayr JA, Zimmermann FA, Fauth C, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet 2011; 89(6): 792-7. doi: 10.1016/j.ajhg.2011.11.011.
pmid: 22152680 |
| 58 |
Okajima K, Korotchkina LG, Prasad C, et al. Mutations of the E1beta subunit gene (PDHB) in four families with pyruvate dehydrogenase deficiency. Mol Genet Metab 2008; 93(4): 371-80. doi: 10.1016/j.ymgme.2007.10.135.
pmid: 18164639 |
| 59 |
Wang G, Ye Q, Ning S, et al. LncRNA MEG3 promotes endoplasmic reticulum stress and suppresses proliferation and invasion of colorectal carcinoma cells through the MEG3/miR-103a-3p/PDHB ceRNA pathway. Neoplasma 2021; 68(2): 362-74. doi: 10.4149/neo_2020_200813N858.
pmid: 33118833 |
| 60 |
Suda C, Yatabe J, Yatabe M, et al. Soluble (pro)renin receptor increased by hypoxia maintains oxidative metabolism in trophoblasts. J Mol Endocrinol 2020; 64(3): 145-54. doi: 10.1530/jme-19-0050.
pmid: 31958319 |
| 61 |
Rowland EA, Snowden CK, Cristea IM. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Curr Opin Chem Biol 2018; 42: 76-85. doi: 10.1016/j.cbpa.2017.11.003.
pmid: 29169048 |
| 62 |
Patel MS, Nemeria NS, Furey W, et al. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem 2014; 289(24): 16615-23. doi: 10.1074/jbc.R114.563148.
pmid: 24798336 |
| 63 |
Anderson NM, Mucka P, Kern JG, et al. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018; 9(2): 216-37. doi: 10.1007/s13238-017-0451-1.
pmid: 28748451 |
| 64 |
Hensley CT, Faubert B, Yuan Q, et al. Metabolic heterogeneity in human lung tumors. Cell 2016; 164(4): 681-94. doi: 10.1016/j.cell.2015.12.034.
pmid: 26853473 |
| 65 | Liu PS, Wang H, Li X, et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017; 18(9): 985-94. doi: 10.1038/ni.3796. |
| 66 |
Hatzivassiliou G, Zhao F, Bauer DE, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 2005; 8(4): 311-21. doi: 10.1016/j.ccr.2005.09.008.
pmid: 16226706 |
| 67 | Zhang X, Wang Y, A G, et al. Pan-cancer analysis of PARP 1 alterations as biomarkers in the prediction of immunotherapeutic effects and the association of its expression levels and immunotherapy signatures. Front Immunol 2021; 12: 721030. doi: 10.3389/fimmu.2021.721030. |
| 68 | Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 2016; 150(7): 1646-58.e17. doi: 10.1053/j.gastro.2016.02.040. |
| 69 |
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013; 19(11): 1423-37. doi: 10.1038/nm.3394.
pmid: 24202395 |
| 70 | Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 2007; 356(2): 125-34. doi: 10.1056/NEJMoa060655. |
| 71 |
Rini BI, Escudier B, Tomczak P, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet 2011; 378(9807): 1931-9. doi: 10.1016/s0140-6736(11)61613-9.
pmid: 22056247 |
| 72 |
Rini BI, Pal SK, Escudier BJ, et al. Tivozanib versus sorafenib in patients with advanced renal cell carcinoma (TIVO-3): a phase 3, multicentre, randomised, controlled, open-label study. Lancet Oncol 2020; 21(1): 95-104. doi: 10.1016/s1470-2045(19)30735-1.
pmid: 31810797 |
| 73 | Lakkakula JR, Gujarathi P, Pansare P, et al. A comprehensive review on alginate-based delivery systems for the delivery of chemotherapeutic agent: Doxorubicin. Carbohydr Polym 2021; 259: 117696. doi: 10.1016/j.carbpol.2021.117696. |
| 74 |
Wei T, Xie XJ, Cao PL. Magnoflorine improves sensitivity to doxorubicin (DOX) of breast cancer cells via inducing apoptosis and autophagy through AKT/mTOR and p38 signaling pathways. Biomed Pharmacother 2020; 121: 109139. doi: 10.1016/j.biopha.2019.109139.
pmid: 31707337 |
| 75 |
Wu Q, Li W, Zhao J, et al. Apigenin ameliorates doxorubicin-induced renal injury via inhibition of oxidative stress and inflammation. Biomed Pharmacother 2021; 137: 111308. doi: 10.1016/j.biopha.2021.111308.
pmid: 33556877 |
| 76 |
Heo JY, Kim HJ, Kim SM, et al. Embelin suppresses STAT3 signaling, proliferation, and survival of multiple myeloma via the protein tyrosine phosphatase PTEN. Cancer Lett 2011; 308(1): 71-80. doi: 10.1016/j.canlet.2011.04.015.
pmid: 21565443 |
| 77 | Lee YJ, Park BS, Park HR, et al. XIAP inhibitor embelin induces autophagic and apoptotic cell death in human oral squamous cell carcinoma cells. Environ Toxicol 2017; 32(11): 2371-78. doi: 10.1002/tox.22450. |
| 78 |
Bisogno G, De Salvo GL, Bergeron C, et al. Vinorelbine and continuous low-dose cyclophosphamide as maintenance chemotherapy in patients with high-risk rhabdomyosarcoma (RMS 2005): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2019; 20(11): 1566-75. doi: 10.1016/s1470-2045(19)30617-5.
pmid: 31562043 |
| 79 | Moskowitz AJ, Shah G, Schöder H, et al. Phase II trial of pembrolizumab plus gemcitabine, vinorelbine, and liposomal doxorubicin as second-line therapy for relapsed or refractory classical Hodgkin lymphoma. J Clin Oncol 2021; 39(28): 3109-17. doi: 10.1200/jco.21.01056. |
| 80 |
Chan A, Verrill M. Capecitabine and vinorelbine in metastatic breast cancer. Eur J Cancer 2009; 45(13): 2253-65. doi: 10.1016/j.ejca.2009.04.031.
pmid: 19464166 |
| 81 |
Kenmotsu H, Yamamoto N, Yamanaka T, et al. Randomized phase III study of pemetrexed plus cisplatin versus vinorelbine plus cisplatin for completely resected stage II to IIIA nonsquamous non-small-cell lung cancer. J Clin Oncol 2020; 38(19): 2187-96. doi: 10.1200/jco.19.02674.
pmid: 32407216 |
| 82 | Falvo P, Orecchioni S, Hillje R, et al. Cyclophosphamide and vinorelbine activate stem-like CD8(+) T cells and improve anti-PD-1 efficacy in triple-negative breast cancer. Cancer Res 2021; 81(3): 685-97. doi: 10.1158/0008-5472.Can-20-1818. |
| 83 | Zhang X, Xiong Y, Xia Q, et al. Efficacy and safety of apatinib plus vinorelbine in patients with wild-type advanced non-small cell lung cancer after second-line treatment failure: a nonrandomized clinical trial. JAMA Netw Open 2020; 3(3): e201226. doi: 10.1001/jamanetworkopen.2020.1226. |
| 84 |
Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018; 15(2): 81-94. doi: 10.1038/nrclinonc.2017.166.
pmid: 29115304 |
| [1] | Wenqin Xu, Jingjing Ye, Tianbing Chen. Identifying and Validating a Novel miRNA-mRNA Regulatory Network Associated with Prognosis in Lung Adenocarcinoma [J]. Chinese Medical Sciences Journal, 2022, 37(1): 31-43. |
| [2] | Li Wenxing, Zhang Yanli. Novel Long Non-coding RNA Markers for Prognostic Prediction of Patients with Bladder Cancer [J]. Chinese Medical Sciences Journal, 2020, 35(3): 239-247. |
| [3] | Zhu Weihua,Xie Wenyong,Zhang Zhedong,Li Shu,Zhang Dafang,Liu Yijun,Zhu Jiye,Leng Xisheng. Postoperative Complications and Survival Analysis of Surgical Resection for Hilar Cholangiocarcinoma: A Retrospective Study of Fifty-Nine Consecutive Patients [J]. Chinese Medical Sciences Journal, 2020, 35(2): 157-169. |
| [4] | Chen Qiang, Zhang Liwei, Huang Dangsheng, Zhang Chunhong, Wang Qiushuang, Shen Dong, Xiong Minjun, Yang Feifei. Five-year Clinical Outcomes of CAD Patients Complicated with Diabetes after StentBoost-optimized Percutaneous Coronary Intervention [J]. Chinese Medical Sciences Journal, 2019, 34(3): 177-183. |
| [5] | Wang Guorong, Wang Zhiwei, Jin Zhengyu. Application and Progress of Texture Analysis in the Therapeutic Effect Prediction and Prognosis of Neoadjuvant Chemoradiotherapy for Colorectal Cancer [J]. Chinese Medical Sciences Journal, 2019, 34(1): 45-50. |
| [6] | Liu Yongsheng, Zhao Yu. Progress in Intraoperative Neurophysiological Monitoring for the Surgical Treatment of Thoracic Spinal Stenosis [J]. Chinese Medical Sciences Journal, 2017, 32(4): 260-264. |
| [7] | Meng-yi Wang, Zhe-yu Niu, Xiang-Gao, Li Zhou, Quan Liao, Yu-pei Zhao. Prognostic Impact of Cell Division Cycle Associated 2 Expression on Pancreatic Ductal Adenocarcinoma [J]. Chinese Medical Sciences Journal, 2016, 31(3): 149-154. |
| [8] | Min Xu, Zheng-song Gu, Cun-zu Wang, Xiao-feng Lu, Ding-chao Xiang, Zhi-cheng Yuan, Qiao-yu Li, Min Wu. Impact of Intraoperative Blood Pressure Control and Temporary Parent Artery Blocking on Prognosis in Cerebral Aneurysms Surgery [J]. Chinese Medical Sciences Journal, 2016, 31(2): 89-94. |
| [9] | Shu-bo Tian, Jian-chun Yu*, Wei-ming Kang, Zhi-qiang Ma, Xin Ye, Chao Yan, Ya-kai Huang. Effect of Neoadjuvant Chemotherapy Treatment on Prognosis of Patients with Advanced Gastric Cancer: a Retrospective Study [J]. Chinese Medical Sciences Journal, 2015, 30(2): 84-89. |
| [10] | Xiao-dong Qu, Resha Shrestha and Mao-de Wang* . Risk Factors Analysis on Traumatic Brain Injury Prognosis [J]. Chinese Medical Sciences Journal, 2011, 26(2): 98-102. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
|