Chinese Medical Sciences Journal ›› 2020, Vol. 35 ›› Issue (4): 306-314.doi: 10.24920/003770
CT纹理分析:结直肠癌KRAS基因突变状态评估的潜在生物标志物
- 北京协和医院放射科,中国医学科学院 北京协和医学院,北京 100730,中国
-
收稿日期:2020-04-30接受日期:2020-07-16出版日期:2020-12-31发布日期:2021-01-08 -
通讯作者:王志伟 E-mail:zhiweiwang1981@sina.com
CT Texture Analysis: A Potential Biomarker for Evaluating KRAS Mutational Status in Colorectal Cancer
Jian Cao,Guorong Wang,Zhiwei Wang( ),Zhengyu Jin
),Zhengyu Jin
- Department of Radiology, Peking Union Medical College Hospital,Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, China
-
Received:2020-04-30Accepted:2020-07-16Published:2020-12-31Online:2021-01-08 -
Contact:Zhiwei Wang E-mail:zhiweiwang1981@sina.com
摘要:
目的 纹理分析可以反映肉眼难以察觉到的肿瘤内异质性。本研究旨在评估CT纹理分析技术鉴别结直肠癌患者KRAS基因突变状态的可行性。
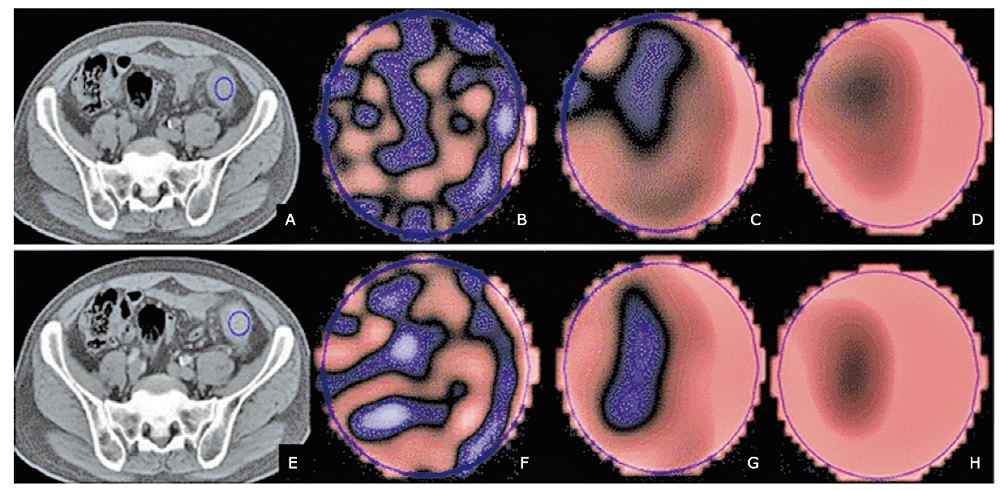
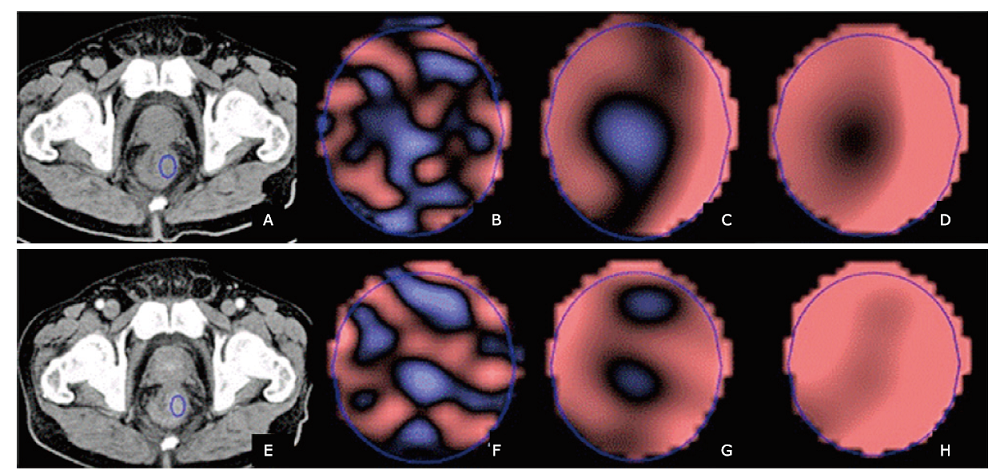
方法 回顾性纳入92例经病理证实且术前接受了腹部增强CT检查的结直肠癌患者。将患者分为训练集(n=51)和验证集(n=41)。使用TexRad纹理分析软件在选定的轴位CT图像上肿瘤区域放置感兴趣区,获得基于不同空间缩放因子(spatial scaling factor, SSF)的特征性纹理参数值,包括均值(mean)、标准差(SD)、熵值(entropy)、偏度值(skewness)、峰值(kurtosis)和正像素均值(mean of positive pixels, MPP)。比较训练集和验证集KRAS突变型患者和野生型患者的CT图像纹理参数值和临床特征(年龄,性别,肿瘤位置,组织病理学,肿瘤大小,TNM分期)。使用皮尔森相关系数计算所有纹理特征的相关性。若某两个纹理特征具有显着的相关性,则删掉较低曲线下面积(area under the curve, AUC)的特征值;纳入最具区分性的单一特征参数,并组合以训练多特征的支持向量机分类器。使用受试者工作特征曲线(receiver operating characteristic, ROC)法评估纹理参数对鉴别结直肠癌患者KRAS突变型与野生型的诊断效能。
结果 在训练集和验证集中,KRAS突变型与野生型患者两组之间的临床特征均无显着差异(P>0.05)。预测结直肠癌患者KRAS突变的最佳模型包括6个纹理特征值,分别是平扫CT中SSF 5的偏度值、SSF 2的熵值、SSF 0的偏度值和峰度值,以及增强CT中SSF3的峰度值和均值。以此建立的诊断模型在训练集中诊断患者KRAS基因突变的曲线下面积为0.951(95% CI:0.895~1,P<0.001),当阈值为0.46时,诊断灵敏度为88.9%,特异度为91.7%。应用于验证集中的曲线下面积为0.995(95% CI:0.982~1, P<0.001),当阈值为0.28时,诊断KRAS突变的灵敏度和特异度分别为100%和93.7%。
结论 利用CT纹理分析技术评估结直肠癌患者KRAS突变状态是可行的。
引用本文
Jian Cao, Guorong Wang, Zhiwei Wang, Zhengyu Jin. CT Texture Analysis: A Potential Biomarker for Evaluating KRAS Mutational Status in Colorectal Cancer[J].Chinese Medical Sciences Journal, 2020, 35(4): 306-314.
"
| Characteristics | Training cohort | Validation cohort | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mutated group (n=27) | Wild-type group (n=24) | t/x2 | P | Mutated group (n=25) | Wild-type group (n=16) | t/x2 | P | ||
| Age (years) | 61.1±10.4 | 59.4±13.9 | 0.258 | 0.620 | 58.0±14.4 | 59.9±9.8 | 0.223 | 0.610 | |
| Gender [n (%)] | 1.457 | 0.227 | 0.009 | 0.923 | |||||
| Male | 9 (33.3) | 12 (50.0) | 9 (36.0) | 6 (37.5) | |||||
| Female | 18 (66.7) | 12 (50.0) | 16 (64.0) | 10 (62.5) | |||||
| Tumor location [n (%)] | 0.695 | 0.952 | 9.163 | 0.057 | |||||
| Ascending colon | 8 (29.6) | 7 (29.2) | 10 (40.0) | 1 (6.3) | |||||
| Transverse colon | 2 (7.4) | 1 (4.2) | 0 (0) | 1 (6.3) | |||||
| Descending colon | 2 (7.4) | 1 (4.2) | 0 (0) | 2 (12.5) | |||||
| Sigmoid colon | 6 (22.2) | 7 (29.2) | 5 (20.0) | 4 (25.0) | |||||
| Rectum | 9 (33.3) | 8 33.3) | 10 (40.0) | 8 (50.0) | |||||
| Tumor size (mm) | 14.6±3.3 | 15.2±3.8 | 0.357 | 0.553 | 15.4±3.9 | 14.2±3.4 | 0.898 | 0.349 | |
| Histological grade [n (%)] | 1.153 | 0.562 | 0.391 | 0.822 | |||||
| Well | 8 (29.6) | 5 (20.8) | 3 (12.0) | 2 (12.5) | |||||
| Moderate | 14 (51.9) | 16 (66.7) | 17 (68.0) | 12 (75.0) | |||||
| Poor | 5 (18.5) | 3 (12.5) | 5 (20.0) | 2 (12.5) | |||||
| T stage [n (%)] | 1.979 | 0.372 | 1.128 | 0.288 | |||||
| T1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||||
| T2 | 4 (14.8) | 1 (4.2) | 0 (0) | 0 (0) | |||||
| T3 | 12 (44.4) | 10 (41.7) | 13 (52.0) | 11 (68.8) | |||||
| T4 | 11 (40.7) | 13 (54.2) | 12 (48.0) | 5 (31.3) | |||||
| N stage [n (%)] | 2.204 | 0.332 | 2.046 | 0.360 | |||||
| N0 | 8 (29.6) | 3 (12.5) | 9 (36.0) | 4 (25.0) | |||||
| N1 | 9 (33.3) | 10 (41.7) | 10 (40.0) | 10 (62.5) | |||||
| N2 | 10 (37.0) | 11 (45.8) | 6 (24.0) | 2 (12.5) | |||||
| M stage [n (%)] | 2.422 | 0.120 | 0.010 | 0.922 | |||||
| M0 | 4 (14.8) | 8 (33.3) | 5 (20.0) | 3 (18.8) | |||||
| M1 | 23 (85.2) | 16 (66.7) | 20 (80.0) | 13 (81.3) | |||||
"
| Models | CT images | Selected features | Training cohort (n=51) | Validation cohort (n=41) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Cut-off value | Sen (%) | Spe (%) | AUC (95% CI) | Cut-off value | Sen (%) | Spe (%) | ||||
| Model 1 | CE | entropy (CT_SSF 2) | 0.951 (0.895-1) | 4.11* | 91.7 | 88.9 | 0.951 (0.891-1) | 4.06* | 100 | 84 | |
| Model 2 | Non-CE | skewness (CT_SSF 5) | 0.951 (0.895-1) | 0.46# | 88.9 | 91.7 | 0.995 (0.982-1) | 0.28# | 100 | 93.7 | |
| CE | skewness (CT_SSF 0) | ||||||||||
| CE | entropy (CT_SSF 2) | ||||||||||
| CE | kurtosis (CT_SSF 0) | ||||||||||
| CE | kurtosis (CT_SSF 3) | ||||||||||
| CE | mean (CT_SSF 3) | ||||||||||
"
| Models | CT images | Selected features | Training cohort | Validation cohort | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Cut-off value | Sen (%) | Spe (%) | AUC (95% CI) | Cut-off value | Sen (%) | Spe (%) | ||||
| Model 3 | Non-CE | MPP (CT_SSF 0) entropy (CT_SSF 2) skewness (CT_SSF 3) kurtosis (CT_SSF 5) | 0.975 (0.939-1) | 0.39* | 96.3 | 91.7 | 0.963 (0.907-1) | 0.79* | 88.0 | 93.7 | |
| Model 4 | CE | kurtosis (CT_SSF 0) entropy (CT_SSF 2) kurtosis (CT_SSF 3) skewness (CT_SSF 4) | 0.951 (0.895-1) | 0.46* | 88.9 | 91.7 | 0.951 (0.891-1) | 0.80* | 84.0 | 100 | |
| 1. |
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68(6):394-424. doi: 10.3322/caac.21492.
doi: 10.3322/caac.21492 pmid: 30207593 |
| 2. |
Freeman HJ. Heterogeneity of colorectal adenomas, the serrated adenoma, and implications for screening and surveillance. World J Gastroenterol 2008; 14(22):3461-3. doi: 10.3748/wjg.14.3461.
doi: 10.3748/wjg.14.3461 pmid: 18567071 |
| 3. | Yu GZ, Chen Y, Long YQ, et al. New insight into the key proteins and pathways involved in the metastasis of colorectal carcinoma. Oncol reports 2008; 19(5):1191-204. doi: 10.3892/or.19.5.1191. |
| 4. |
Labianca R, Nordlinger B, Beretta GD, et al. Primary colon cancer: ESMO Clinical Practice Guidelines for diagnosis, adjuvant treatment and follow-up. Ann Oncol 2010; 21(Suppl 5):v70-7. doi: 10.1093/annonc/mdq168.
doi: 10.1093/annonc/mdq168 |
| 5. |
Ulivi P, Capelli L, Valgiusti M, et al. Predictive role of multiple gene alterations in response to cetuximab in metastatic colorectal cancer: a single center study. J Translation Med 2012; 10:87. doi: 10.1186/1479-5876-10-87.
doi: 10.1186/1479-5876-10-87 |
| 6. |
Berger MD, Stintzing S, Heinemann V, et al. Impact of genetic variations in the MAPK signaling pathway on outcome in metastatic colorectal cancer patients treated with first-line FOLFIRI and bevacizumab: data from FIRE-3 and TRIBE trials. Ann Oncol 2017; 28(11):2780-85. doi: 10.1093/annonc/mdx412.
pmid: 29045529 |
| 7. | Baselga J. The EGFR as a target for anticancer therapy:focus on cetuximab. Eur J Cancer 2001; 37(Suppl 4):S16-22. doi: 10.1016/s0959-8049(01)00233-7. |
| 8. |
Napolitano S, Martini G, Martinelli E, et al. Therapeutic efficacy of SYM004, a mixture of two anti-EGFR antibodies in human colorectal cancer with acquired resistance to cetuximab and MET activation. Oncotarget 2017; 8(40):67592-604. doi: 10.18632/oncotarget.18749.
doi: 10.18632/oncotarget.18749 pmid: 28978055 |
| 9. |
Cremolini C, Schirripa M, Antoniotti C, et al. First-line chemotherapy for mCRC-a review and evidence-based algorithm. Nature Reviews Clin Oncol 2015; 12(10):607-19. doi: 10.1038/nrclinonc.2015.129.
doi: 10.1038/nrclinonc.2015.129 |
| 10. |
Michl M, Stintzing S, Fischer von Weikersthal L, et al. CEA response is associated with tumor response and survival in patients with KRAS exon 2 wild-type and extended RAS wild-type metastatic colorectal cancer receiving first-line FOLFIRI plus cetuximab or bevacizumab (FIRE-3 trial). Ann Oncol 2016; 27(8):1565-72. doi: 10.1093/annonc/mdw222.
pmid: 27234640 |
| 11. |
Tsujikawa T, Yamamoto M, Shono K, et al. Assessment of intratumor heterogeneity in mesenchymal uterine tumor by an (18)F-FDG PET/CT texture analysis. Ann Nucl Med 2017; 31(10):752-57. doi: 10.1007/s12149-017-1208-x.
pmid: 28905201 |
| 12. | Zhang GM, Shi B, Sun H, et al. Differentiating pheochromocytoma from lipid-poor adrenocortical adenoma by CT texture analysis: feasibility study. Abd Radiol 2017; 42(9):2305-13. doi: 10.1007/s00261-017-1118-3. |
| 13. |
Liu S, Zheng H, Pan X, et al. Texture analysis of CT imaging for assessment of esophageal squamous cancer aggressiveness. J Thorac Dis 2017; 9(11):4724-32. doi: 10.21037/jtd.2017.06.46.
pmid: 29268543 |
| 14. |
Liu Y, Liu S, Qu F, et al. Tumor heterogeneity assessed by texture analysis on contrast-enhanced CT in lung adenocarcinoma: association with pathologic grade. Oncotarget 2017; 8(32):53664-74. doi: 10.18632/oncotarget.15399.
doi: 10.18632/oncotarget.15399 pmid: 28881840 |
| 15. |
Kim HS, Kim JH, Yoon YC, et al. Tumor spatial heterogeneity in myxoid-containing soft tissue using texture analysis of diffusion-weighted MRI. PLoS One 2017; 12(7):e0181339. doi: 10.1371/journal.pone.0181339.
pmid: 28708850 |
| 16. |
Craigie M, Squires J, Miles K. Can CT measures of tumour heterogeneity stratify risk for nodal metastasis in patients with non-small cell lung cancer? Clin Radiol 2017; 72(10):899.e1-9899.e7. doi: 10.1016/j.crad.2017.04.013.
doi: 10.1016/j.crad.2017.04.013 |
| 17. |
Miles KA, Ganeshan B, Rodriguez-Justo M, et al. Multifunctional imaging signature for V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in colorectal cancer. J Nucl Med 2014; 55(3):386-91. doi: 10.2967/jnumed.113.120485.
doi: 10.2967/jnumed.113.120485 |
| 18. |
Ukweh ON, Ugbem TI, Okeke CM, et al. Value and diagnostic efficacy of fetal morphology assessment using ultrasound in a poor-resource setting. Diagnost 2019; 9(3):109. doi: 10.3390/diagnostics9030109.
doi: 10.3390/diagnostics9030109 |
| 19. |
Feng C, Lu F, Shen Y, et al. Tumor heterogeneity in gastrointestinal stromal tumors of the small bowel: volumetric CT texture analysis as a potential biomarker for risk stratification. Cancer imaging 2018; 18(1):46. doi: 10.1186/s40644-018-0182-4.
doi: 10.1186/s40644-018-0182-4 pmid: 30518436 |
| 20. |
Lubner MG, Smith AD, Sandrasegaran K, et al. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 2017; 37(5):1483-503. doi: 10.1148/rg.2017170056.
doi: 10.1148/rg.2017170056 pmid: 28898189 |
| 21. |
Weiss GJ, Ganeshan B, Miles KA, et al. Noninvasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS One 2014; 9(7):e100244. doi: 10.1371/journal.pone.0100244.
doi: 10.1371/journal.pone.0100244 pmid: 24987838 |
| 22. |
Ba-Ssalamah A, Muin D, Schernthaner R, et al. Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur J Radiol 2013; 82(10):e537-43. doi: 10.1016/j.ejrad.2013.06.024.
doi: 10.1016/j.ejrad.2013.06.024 |
| 23. | Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abd Imaging 2015; 40(7):2331-7. doi: 10.1007/s00261-015-0438-4. |
| 24. |
Beckers RCJ, Trebeschi S, Maas M, et al. CT texture analysis in colorectal liver metastases and the surrounding liver parenchyma and its potential as an imaging biomarker of disease aggressiveness, response and survival. Eur J Radiol 2018; 102:15-21. doi: 10.1016/j.ejrad.2018.02.031.
pmid: 29685529 |
| 25. |
Lee SJ, Zea R, Kim DH, et al. CT texture features of liver parenchyma for predicting development of metastatic disease and overall survival in patients with colorectal cancer. Eur Radiol 2018; 28(4):1520-28. doi: 10.1007/s00330-017-5111-6.
doi: 10.1007/s00330-017-5111-6 pmid: 29164382 |
| 26. |
Chee CG, Kim YH, Lee KH, et al. CT texture analysis in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy: A potential imaging biomarker for treatment response and prognosis. PLoS One 2017; 12(8):e0182883. doi: 10.1371/journal.pone.0182883.
doi: 10.1371/journal.pone.0182883 pmid: 28797063 |
| 27. |
Lovinfosse P, Koopmansch B, Lambert F, et al. (18)F-FDG PET/CT imaging in rectal cancer: relationship with the RAS mutational status. Br J Radiol 2016; 89(1063):20160212. doi: 10.1259/bjr.20160212.
doi: 10.1259/bjr.20160212 pmid: 27146067 |
| 28. |
Miles KA, Ganeshan B, Rodriguez-Justo M, et al. Multifunctional imaging signature for V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in colorectal cancer. J Nucl Med 2014; 55(3):386-91. doi: 10.2967/jnumed.113.120485.
doi: 10.2967/jnumed.113.120485 pmid: 24516257 |
| 29. |
Chen SW, Chiang HC, Chen WT, et al. Correlation between PET/CT parameters and KRAS expression in colorectal cancer. Clin Nucl Med 2014; 39(8):685-9. doi: 10.1097/rlu.0000000000000481.
pmid: 24978328 |
| 30. |
Kawada K, Nakamoto Y, Kawada M, et al. Relationship between 18F-fluorodeoxyglucose accumulation and KRAS/BRAF mutations in colorectal cancer. Clin Cancer Res 2012; 18(6):1696-703. doi: 10.1158/1078-0432.ccr-11-1909.
pmid: 22282467 |
| 31. |
Kawada K, Toda K, Nakamoto Y, et al. Relationship between 18F-FDG PET/CT scans and KRAS mutations in metastatic colorectal cancer. J Nucl Med 2015; 56(9):1322-7. doi: 10.2967/jnumed.115.160614.
doi: 10.2967/jnumed.115.160614 pmid: 26135109 |
| 32. |
Yang L, Dong D, Fang M, et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol 2018; 28(5):2058-67. doi: 10.1007/s00330-017-5146-8.
doi: 10.1007/s00330-017-5146-8 pmid: 29335867 |
| [1] | 杨威, 张梅. 抗癌治疗中对心脏毒性效应具有潜在预测价值的生物标志物[J]. Chinese Medical Sciences Journal, 2021, 36(4): 333-341. |
| [2] | 王站, 王旭, 王文达, 郑国洋, 郭浩, 张玉石. 术前中性粒细胞与淋巴细胞比值预测可切除泌尿系肿瘤预后的价值:系统综述和荟萃分析[J]. Chinese Medical Sciences Journal, 2020, 35(3): 262-271. |
| [3] | 吴建强, 秦伟伟, 潘利, 王小蓉, 张彪, 单广良, 高友鹤. 中国不同地区健康人尿液蛋白质组的地域差异[J]. Chinese Medical Sciences Journal, 2019, 34(3): 157-167. |
| [4] | 王英伟, 张兴华, 王波涛, 王叶, 刘梦琦, 王海屹, 叶慧义. 体素内不相干运动成像参数的纹理分析在胰腺神经内分泌肿瘤和胰腺癌鉴别诊断中的价值[J]. Chinese Medical Sciences Journal, 2019, 34(1): 1-9. |
| [5] | 王波涛, 刘明霞, 陈志晔. 磁共振T2加权成像纹理特征分析在脑胶质母细胞瘤与脑原发性中枢神经系统淋巴瘤鉴别诊断中的价值[J]. Chinese Medical Sciences Journal, 2019, 34(1): 10-17. |
| [6] | 刘洪娟, 周欢粉, 宗林雄, 刘梦琦, 魏世辉, 陈志晔. 视神经炎患者视神经磁共振成像直方图分析[J]. Chinese Medical Sciences Journal, 2019, 34(1): 18-23. |
| [7] | 徐佳, 王萱, 金征宇, 游燕, 王勤, 王士阗, 薛华丹. 钆塞酸二钠增强磁共振图像纹理分析对于评价大鼠肝纤维化的价值[J]. Chinese Medical Sciences Journal, 2019, 34(1): 24-32. |
| [8] | 王波涛, 樊文萍, 许欢, 李丽慧, 张晓欢, 王昆, 刘梦琦, 游俊浩, 陈志晔. 磁共振扩散加权成像纹理特征分析在乳腺良恶性肿瘤鉴别中的价值[J]. Chinese Medical Sciences Journal, 2019, 34(1): 33-37. |
| [9] | 王国蓉, 王志伟, 金征宇. 纹理分析在结直肠癌新辅助放化疗疗效预测及预后分析中的应用及研究进展[J]. Chinese Medical Sciences Journal, 2019, 34(1): 45-50. |
| [10] | 李涛, 杨立, 张卫国, 罗春才, 黄自立, 李金锋, 李欣. 6 4层多排CT对冠状动脉旁路移植术后的中期随访:影响桥血管通畅性的危险因素研究[J]. Chinese Medical Sciences Journal, 2018, 33(2): 69-76. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
|