Chinese Medical Sciences Journal ›› 2021, Vol. 36 ›› Issue (3): 204-209.doi: 10.24920/003962
Special Issue: 人工智能与精准肿瘤学
• Original Article • Previous Articles Next Articles
Histopathological Diagnosis System for Gastritis Using Deep Learning Algorithm
Wei Ba1, Shuhao Wang2, Cancheng Liu2, Yuefeng Wang2, Huaiyin Shi1, Zhigang Song1, *( )
)
- 1Department of Pathology, Chinese PLA General Hospital & Medical School, Beijing 100853, China
2Artificial Intelligence Lab, Thorough Images,Beijing 100853, China
-
Received:2021-06-29Accepted:2021-08-18Published:2021-09-30Online:2021-08-31 -
Contact:Zhigang Song E-mail:songzhg301@139.com
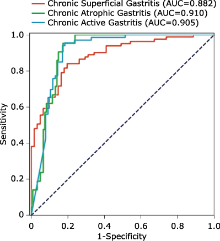
| A deep learning algorithm for pathological classification of chronic superficial, active, atrophic gastritis was developed and assessed using whole-slide images (WSIs), which demonstrated good performance. |
Cite this article
Wei Ba, Shuhao Wang, Cancheng Liu, Yuefeng Wang, Huaiyin Shi, Zhigang Song. Histopathological Diagnosis System for Gastritis Using Deep Learning Algorithm[J].Chinese Medical Sciences Journal, 2021, 36(3): 204-209.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Pathological characteristics of the whole-slide images datasets for gastritis WSIs (n)"
| Datasets | Normal mucosa | Chronic superficial gastritis | Chronic atrophic gastritis | Chronic active gastritis | Total |
|---|---|---|---|---|---|
| Training set | 101 | 671 | 83 | 153 | 1008 |
| Validation set | 10 | 29 | 15 | 46 | 100 |
| Test set | 11 | 81 | 21 | 29 | 142 |
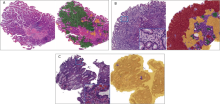
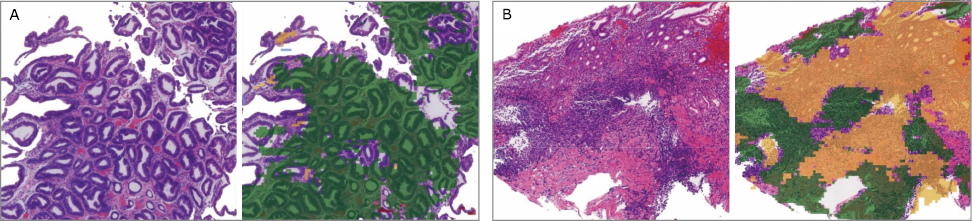
Figure 2.
Examples of chronic atrophic gastritis (CAtG), chronic superficial gastritis (CSuG) and chronic active gastritis (CAcG) in test data set that were correctly diagnosed by the deep learning algorithm (A) CAtG. (B) CSuG. (C) CAcG. Images on the left represent typical histological findings for the disease diagnosis. The green, red, and yellow in the images on the right represent the predicted CAtG, CSuG, and CAcG regions, respectively. The boxes in the lower right corner of (B) and (C) images represent the local magnification of the corresponding pane in the image, which show the infiltrating inflammatory cells of different gastritis."

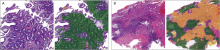
Figure 3.
Examples of chronic superficial gastritis (CSuG) and chronic atrophic gastritis (CAtG) that were incorrectly diagnosed by the deep learning algorithm (A) CSuG was misdiagnosed as CAtG. (B) CAtG was misdiagnosed as chronic active gastritis (CAcG). Images on the right are the outputs of the deep learning algorithm. The green and yellow colors represent the predicted CAtG and CAcG regions by the algorithm."
| 1. |
Correa P, Yardley JH. Grading and classification of chronic gastritis: one American response to the Sydney system. Gastroenterology 1992; 102(1):355-9. doi: 10.1016/0016-5085(92)91820-t.
doi: 10.1016/0016-5085(92)91820-t pmid: 1727769 |
| 2. |
Price AB, Misiewicz JJ. Grading and classification of chronic gastritis: the response of the working party. Gastroenterology 1992; 103(3):1116-7. doi: 10.1016/0016-5085(92)90066-8.
doi: 10.1016/0016-5085(92)90066-8 pmid: 1499919 |
| 3. |
Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 1996; 20(10):1161-81. doi: 10.1097/00000478-199610000-00001.
doi: 10.1097/00000478-199610000-00001 pmid: 8827022 |
| 4. |
Stolte M, Meining A. The updated Sydney system: classification and grading of gastritis as the basis of diagnosis and treatment. Can J Gastroenterol 2001; 15(9):591-8. doi: 10.1155/2001/367832.
doi: 10.1155/2001/367832 |
| 5. |
Metter DM, Colgan TJ, Leung ST, et al. Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw Open 2019; 2(5):e194337. doi: 10.1001/jamanetworkopen.2019.4337.
doi: 10.1001/jamanetworkopen.2019.4337 |
| 6. |
Retamero JA, Aneiros-Fernandez J, Del Moral RG. Complete digital pathology for routine histopathology diagnosis in a multicenter hospital network. Arch Pathol Lab Med 2020; 144(2):221-8. doi: 10.5858/arpa.2018-0541-OA.
doi: 10.5858/arpa.2018-0541-OA pmid: 31295015 |
| 7. |
Cruz-Roa A, Gilmore H, Basavanhally A, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Sci Rep 2017; 7:46450. doi: 10.1038/srep46450.
doi: 10.1038/srep46450 pmid: 28418027 |
| 8. |
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318(2):2199-210. doi: 10.1001/jama.2017.14585.
doi: 10.1001/jama.2017.14585 |
| 9. |
Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nature Medicine 2018; 24(10):1559. doi: 10.1038/s41591-018-0177-5.
doi: 10.1038/s41591-018-0177-5 |
| 10. |
Khosravi P, Kazemi E, Imielinski M, et al. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 2018; 27:317-28. doi: 10.1016/j.ebiom.2017.12.026.
doi: S2352-3964(17)30507-8 pmid: 29292031 |
| 11. |
Ba W, Wang R, Yin G, et al. Diagnostic assessment of deep learning for melanocytic lesions using whole-slide pathological images. Transl Onc 2021; 6(1):475.doi: 10.1016/j.tranon.2021.101161.
doi: 10.1016/j.tranon.2021.101161 |
| 12. |
Golden JA. Deep learning algorithms for detection of lymph node metastases from breast cancer: helping artificial intelligence be seen. JAMA 2017; 318(22):2184-6. doi: 10.1001/jama.2017.14580.
doi: 10.1001/jama.2017.14580 |
| 13. |
Pan Y, Sun Z, Wang W, et al. Automatic detection of squamous cell carcinoma metastasis in esophageal lymph nodes using semantic segmentation. Clin Transl Med 2020; 10(3):e129. doi: 10.1002/ctm2.129.
doi: 10.1002/ctm2.129 |
| 14. |
Martin DR, Hanson JA, Gullapalli RR, et al. A deep learning convolutional neural network can recognize common patterns of injury in gastric pathology. Arch Pathol Lab Med 2020; 144(3):370-8. doi: 10.5858/arpa.2019-0004-OA.
doi: 10.5858/arpa.2019-0004-OA pmid: 31246112 |
| 15. |
Song Z, Zou S, Zhou W, et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun 2020; 11(1):4294. doi: 10.1038/s41467-020-18147-8.
doi: 10.1038/s41467-020-18147-8 |
| 16. |
Arvaniti E, Fricker KS, Moret M, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 2018; 8(1):12054. doi: 10.1038/s41598-018-30535-1.
doi: 10.1038/s41598-018-30535-1 pmid: 30104757 |
| 17. |
Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25(1):44-56. doi: 10.1038/s41591-018-0300-7.
doi: 10.1038/s41591-018-0300-7 pmid: 30617339 |
| 18. |
Tschandl P. Problems and potentials of automated object detection for skin cancer recognition. JAMA Dermatol 2020; 156(1):23-4. doi: 10.1001/jamadermatol.2019.3360.
doi: 10.1001/jamadermatol.2019.3360 pmid: 31799989 |
| [1] | Runnan Cao, Mengjie Fang, Hailing Li, Jie Tian, Di Dong. Semi-supervised Long-tail Endoscopic Image Classification [J]. Chinese Medical Sciences Journal, 2022, 37(3): 171-180. |
| [2] | Ying Liu, Yongjun Ma, Caiqun Huang. Evaluation of the Gastric Microbiome in Patients with Chronic Superficial Gastritis and Intestinal Metaplasia [J]. Chinese Medical Sciences Journal, 2022, 37(1): 44-51. |
| [3] | Chen Xu, Huo Xiaofei, Wu Zhe, Lu Jingjing. Advances of Artificial Intelligence Application in Medical Imaging of Ovarian Cancers [J]. Chinese Medical Sciences Journal, 2021, 36(3): 196-203. |
| [4] | Lianyan Xu, Ke Yan, Le Lu, Weihong Zhang, Xu Chen, Xiaofei Huo, Jingjing Lu. External and Internal Validation of a Computer Assisted Diagnostic Model for Detecting Multi-Organ Mass Lesions in CT images [J]. Chinese Medical Sciences Journal, 2021, 36(3): 210-217. |
| [5] | Jiazheng Li, Lei Tang. Radiomics in Antineoplastic Agents Development: Application and Challenge in Response Evaluation [J]. Chinese Medical Sciences Journal, 2021, 36(3): 187-195. |
| [6] | Dasheng Li,Dawei Wang,Nana Wang,Haiwang Xu,He Huang,Jianping Dong,Chen Xia. An Insight of the First Community Infected COVID-19 Patient in Beijing by Imported Case: Role of Deep Learning-Assisted CT Diagnosis [J]. Chinese Medical Sciences Journal, 2021, 36(1): 66-71. |
| [7] | Wang Zheng, Zhao Qinghua, Yang Jinglin, Zhou Feng. Enhancing Quality of Patients Care and Improving Patient Experience in China with Assistance of Artificial Intelligence [J]. Chinese Medical Sciences Journal, 2020, 35(3): 286-288. |
| [8] | Yang Xiaolin, Wang Zhe, Pan Hongjie, Zhu Yan. Ontology: Footstone for Strong Artificial Intelligence [J]. Chinese Medical Sciences Journal, 2019, 34(4): 277-280. |
| [9] | Li Peilin, Yuan Zhenming, Tu Wenbo, Yu Kai, Lu Dongxin. Medical Knowledge Extraction and Analysis from Electronic Medical Records Using Deep Learning [J]. Chinese Medical Sciences Journal, 2019, 34(2): 133-139. |
| [10] | Shi Ying-huan,Wang Qian. The Artificial Intelligence-Enabled Medical Imaging: Today and Its Future [J]. Chinese Medical Sciences Journal, 2019, 34(2): 71-75. |
| [11] | Xiao Yi,Liu Shiyuan. Collaborations of Industry, Academia, Research and Application Improve the Healthy Development of Medical Imaging Artificial Intelligence Industry in China [J]. Chinese Medical Sciences Journal, 2019, 34(2): 84-88. |
| [12] | Guan Jian. Artificial Intelligence in Healthcare and Medicine: Promises, Ethical Challenges and Governance [J]. Chinese Medical Sciences Journal, 2019, 34(2): 76-83. |
| [13] | Chinese Innovative Alliance of Industry, Education, Research and Application of Artificial Intelligence for Medical . Releasing of The White Paper on Medical Imaging Artificial Intelligence in China [J]. Chinese Medical Sciences Journal, 2019, 34(2): 89-89. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
|